|
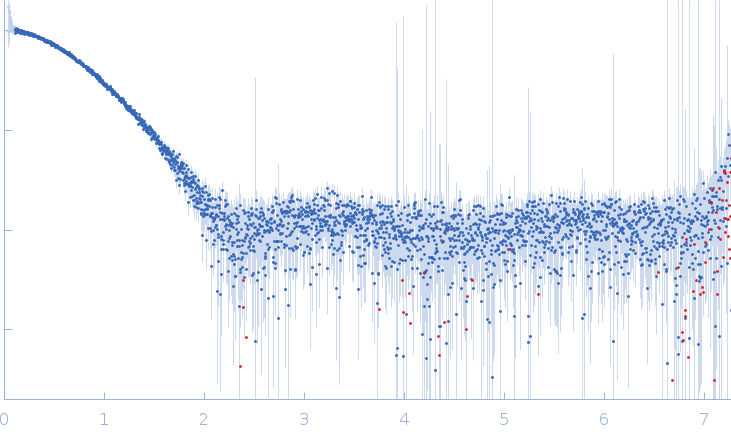
Synchrotron SAXS data from solutions of the nonstructural 2A protein of human parechovirus 1 (HPeV1-2A) in 20 mM HEPES, 150 mM NaCl, 5 mM DDT, pH 7.4 were collected on the EMBL P12 beam line at PETRA III (DESY; Hamburg, Germany) using a Pilatus 6M detector at a sample-detector distance of 3 m and at a wavelength of λ = 0.124 nm (I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle). One solute concentration of 2.84 mg/ml was measured at 10°C. 30 successive 0.050 second frames were collected. The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted.
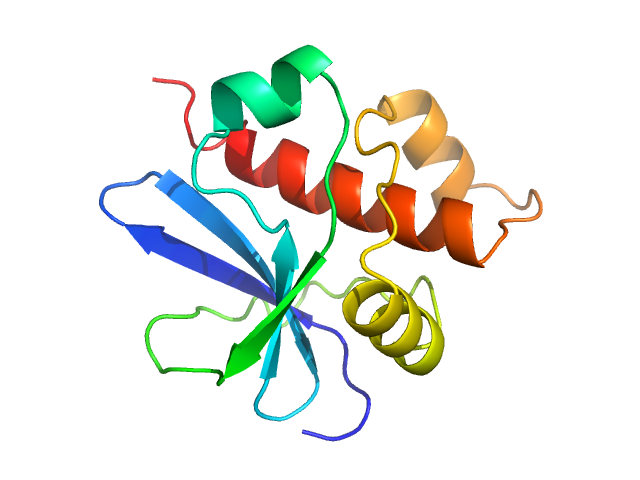
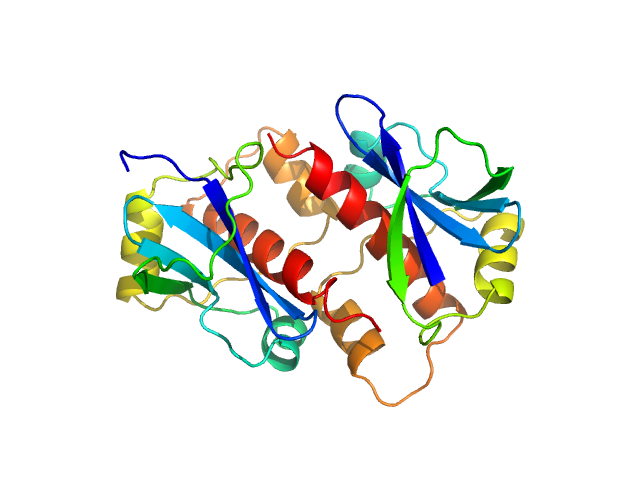
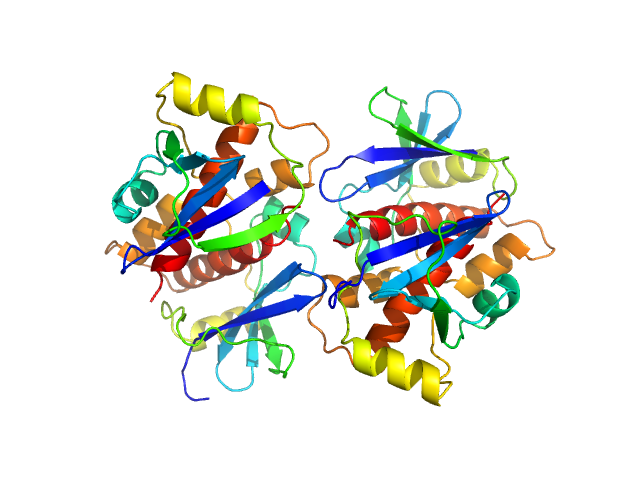
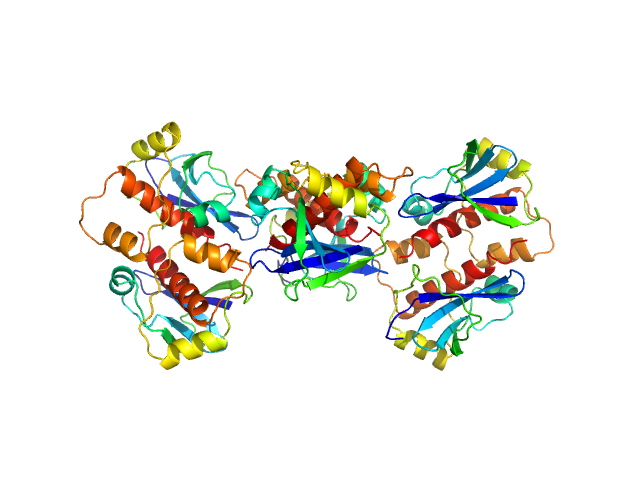
The HPeV1-2A (1-129) sample was measured at various sample concentrations. For this entry the 2.84 mg/ml sample scattering is displayed, while measurements spanning seven dilutions are made available in the full entry zip archive. Mixture analysis of the scattering profiles was performed with the program OLIGOMER. The OLIGOMER results show that HPeV1-2A is a mixture of monomers (74%), dimers (10.6%), tetramers (13%), hexamers (0.24%), based on the volume-fraction weighted form factors calculated from the X-ray crystal structure, PDB ID: 7ZU4 (dimer) and derived monomer, tetramer and hexamer (based on crystal packing).
|
|
 s, nm-1
s, nm-1