|
|
|
|
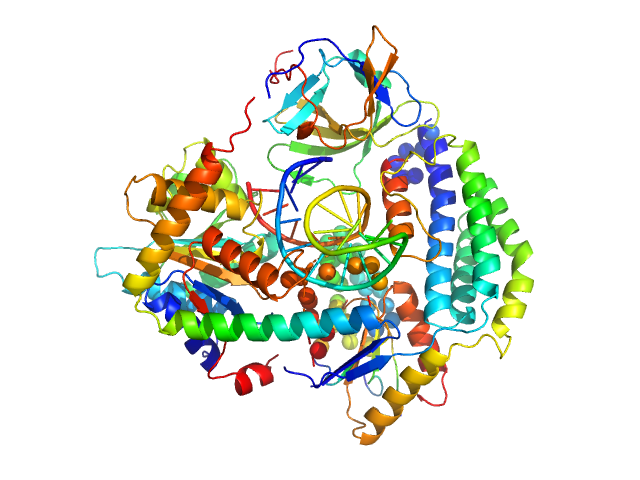
|
| Sample: |
Probable ATP-dependent RNA helicase DDX58 (without CARDs) monomer, 80 kDa Homo sapiens protein
5´ppp 10mer hairpin dsRNA monomer, 8 kDa RNA
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, 2.5 mM MgCl2, 10% glycerol and 1mM DTT, 0.5 mM AMP-PNP, pH: 7.4
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2015 May 29
|
...hairpin RNA in the activation of RIG-I revealed by solution-based analysis.
Nucleic Acids Res 46(6):3169-3186 (2018)
Shah N, Beckham SA, Wilce JA, Wilce MCJ
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
135 |
nm3 |
|