|
|
|
|
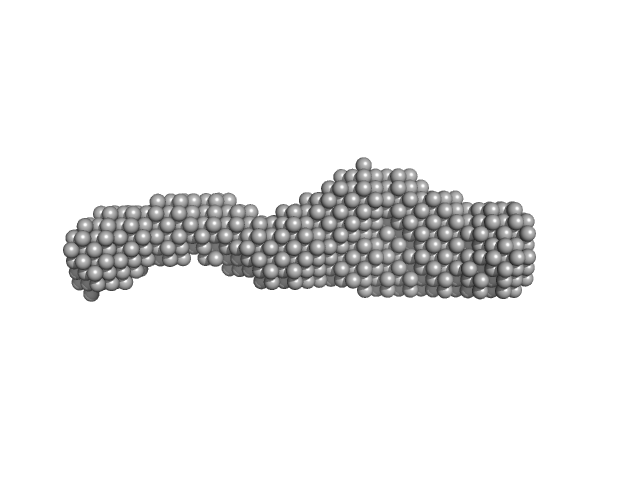
|
| Sample: |
Nucleoporin POM152 monomer, 49 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
10mM HEPES, 150mM NaCl, 10%(v/v) glycerol, 5mM DTT, pH: 7.5
|
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2015 Apr 12
|
Molecular Architecture of the Major Membrane Ring Component of the Nuclear Pore Complex.
Structure 25(3):434-445 (2017)
...Cowburn D, Almo SC, Sali A, Rout MP, Fernandez-Martinez J
|
| RgGuinier |
4.3 |
nm |
| Dmax |
15.4 |
nm |
| VolumePorod |
67 |
nm3 |
|