|
Synchrotron SAXS data from solutions of HIV-1 Primer Binding Site (PBS)-Segment RNA in 10 mM Tris, 140 mM KCl, 10 mM NaCl, 1 mM MgCl2, pH 7.5 were collected on the BioCAT 18ID beam line at the Advanced Photon Source (APS), Argonne National Laboratory storage ring (Lemont, IL, USA) using a Pilatus3 X 1M detector at a wavelength of λ = 0.103 nm (I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle). In-line size-exclusion chromatography (SEC) SAS was employed. The SEC parameters were as follows: A 200.00 μl sample at 9 mg/ml was injected at a 0.75 ml/min flow rate onto a GE Superdex 200 Increase 10/300 column at 25°C. 876 successive 1 second frames were collected every 2 seconds from the outlet stream of the SEC column. The data were normalized to the intensity of the transmitted beam and radially averaged. The buffer scattering curves measured immediately before the elution peak of the sample were used for background correction.
Sample detector distance = UNKNOWN
|
|
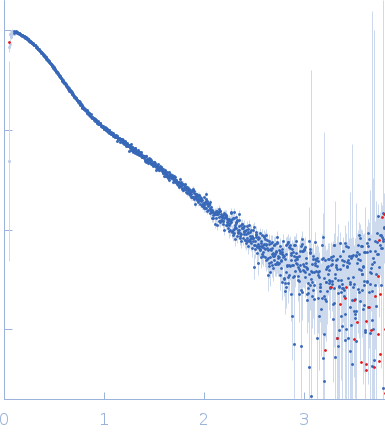 s, nm-1
s, nm-1
