UniProt ID: O08967 (63-399) Cytohesin-3
|
|
|

|
| Sample: |
Cytohesin-3 monomer, 40 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 2 mM MgCl2, 0.1% 2-mercaptoethanol, 5% glycerol, 0.001 mM insitol 1,3,4,5-tetrakis phosphate, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2013 Nov 13
|
Structural Dynamics Control Allosteric Activation of Cytohesin Family Arf GTPase Exchange Factors.
Structure 26(1):106-117.e6 (2018)
Malaby AW, Das S, Chakravarthy S, Irving TC, Bilsel O, Lambright DG
|
| RgGuinier |
2.8 |
nm |
| Dmax |
11.2 |
nm |
| VolumePorod |
61 |
nm3 |
|
|
UniProt ID: O08967 (63-399) Cytohesin-3
|
|
|
|
| Sample: |
Cytohesin-3 monomer, 40 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 2 mM MgCl2, 0.001 mM inositol 1,3,4,5-tetrakisphosphate, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2013 Mar 21
|
Structural Dynamics Control Allosteric Activation of Cytohesin Family Arf GTPase Exchange Factors.
Structure 26(1):106-117.e6 (2018)
Malaby AW, Das S, Chakravarthy S, Irving TC, Bilsel O, Lambright DG
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.3 |
nm |
| VolumePorod |
64 |
nm3 |
|
|
UniProt ID: O08967 (14-399) Cytohesin-3
|
|
|

|
| Sample: |
Cytohesin-3 dimer, 93 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 2 mM MgCl2, 0.1% 2-mercaptoethanol, 5% glycerol, 0.001 mM insitol 1,3,4,5-tetrakis phosphate, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2013 Nov 15
|
Structural Organization and Dynamics of Homodimeric Cytohesin Family Arf GTPase Exchange Factors in Solution and on Membranes.
Structure (2019)
Das S, Malaby AW, Nawrotek A, Zhang W, Zeghouf M, Maslen S, Skehel M, Chakravarthy S, Irving TC, Bilsel O, Cherfils J, Lambright DG
|
| RgGuinier |
5.5 |
nm |
| Dmax |
26.0 |
nm |
| VolumePorod |
194 |
nm3 |
|
|
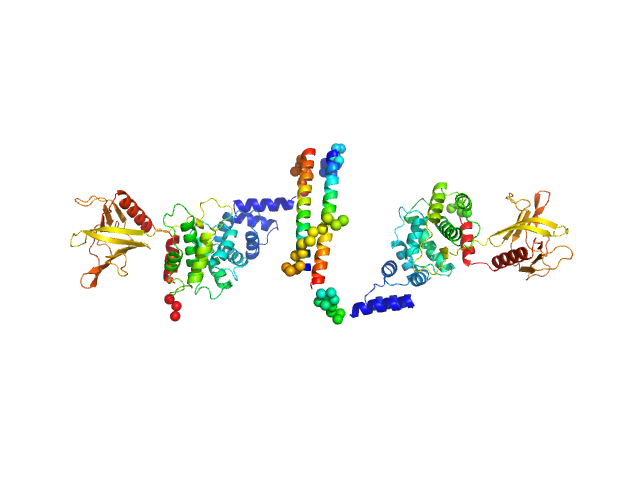
UniProt ID: O08967 (14-399) Cytohesin-3
|
|
|
|
| Sample: |
Cytohesin-3 dimer, 93 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 2 mM MgCl2, 0.1% 2-mercaptoethanol, 5% glycerol, 0.001 mM insitol 1,3,4,5-tetrakis phosphate, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2013 Nov 15
|
Structural Organization and Dynamics of Homodimeric Cytohesin Family Arf GTPase Exchange Factors in Solution and on Membranes.
Structure (2019)
Das S, Malaby AW, Nawrotek A, Zhang W, Zeghouf M, Maslen S, Skehel M, Chakravarthy S, Irving TC, Bilsel O, Cherfils J, Lambright DG
|
| RgGuinier |
5.5 |
nm |
| Dmax |
26.0 |
nm |
| VolumePorod |
194 |
nm3 |
|
|
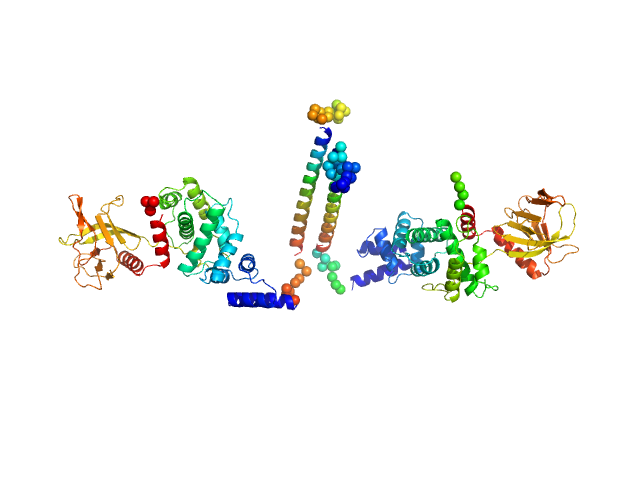
UniProt ID: O08967 (14-399) Cytohesin-3
|
|
|
|
| Sample: |
Cytohesin-3 dimer, 93 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 2 mM MgCl2, 0.1% 2-mercaptoethanol, 5% glycerol, 0.001 mM insitol 1,3,4,5-tetrakis phosphate, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2013 Nov 15
|
Structural Organization and Dynamics of Homodimeric Cytohesin Family Arf GTPase Exchange Factors in Solution and on Membranes.
Structure (2019)
Das S, Malaby AW, Nawrotek A, Zhang W, Zeghouf M, Maslen S, Skehel M, Chakravarthy S, Irving TC, Bilsel O, Cherfils J, Lambright DG
|
| RgGuinier |
5.5 |
nm |
| Dmax |
26.0 |
nm |
| VolumePorod |
194 |
nm3 |
|
|
UniProt ID: O08967 (14-390) Cytohesin-3
|
|
|

|
| Sample: |
Cytohesin-3 dimer, 90 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 2 mM MgCl2, 0.1% 2-mercaptoethanol, 5% glycerol, 0.001 mM insitol 1,3,4,5-tetrakis phosphate, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2013 Nov 15
|
Structural Organization and Dynamics of Homodimeric Cytohesin Family Arf GTPase Exchange Factors in Solution and on Membranes.
Structure (2019)
Das S, Malaby AW, Nawrotek A, Zhang W, Zeghouf M, Maslen S, Skehel M, Chakravarthy S, Irving TC, Bilsel O, Cherfils J, Lambright DG
|
| RgGuinier |
5.3 |
nm |
| Dmax |
25.7 |
nm |
| VolumePorod |
168 |
nm3 |
|
|
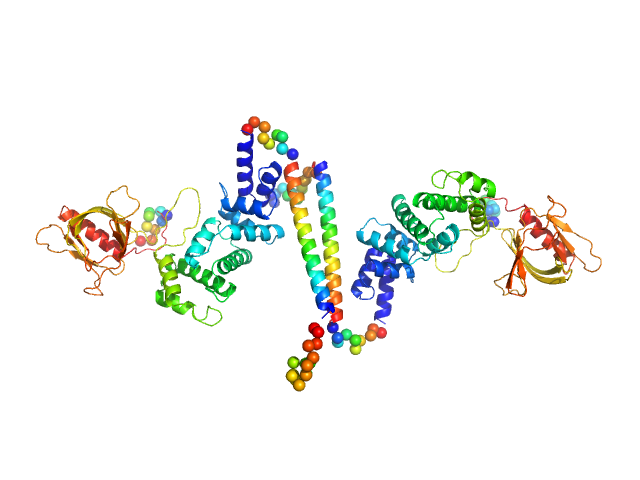
UniProt ID: O08967 (14-390) Cytohesin-3
|
|
|
|
| Sample: |
Cytohesin-3 dimer, 90 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 2 mM MgCl2, 0.1% 2-mercaptoethanol, 5% glycerol, 0.001 mM insitol 1,3,4,5-tetrakis phosphate, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2013 Nov 15
|
Structural Organization and Dynamics of Homodimeric Cytohesin Family Arf GTPase Exchange Factors in Solution and on Membranes.
Structure (2019)
Das S, Malaby AW, Nawrotek A, Zhang W, Zeghouf M, Maslen S, Skehel M, Chakravarthy S, Irving TC, Bilsel O, Cherfils J, Lambright DG
|
| RgGuinier |
5.1 |
nm |
| Dmax |
25.7 |
nm |
| VolumePorod |
168 |
nm3 |
|
|
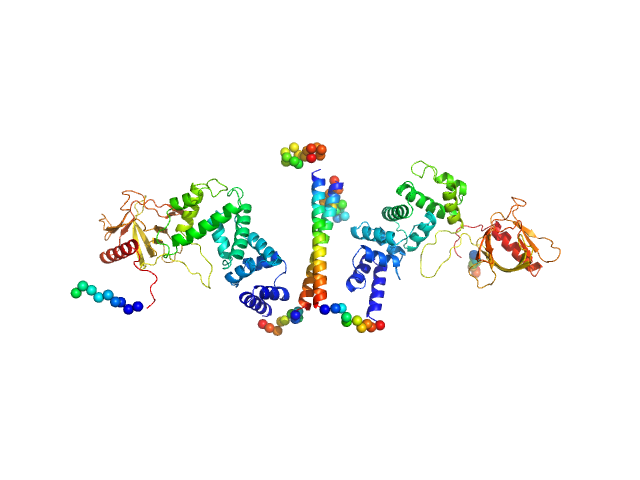
UniProt ID: O08967 (14-390) Cytohesin-3
|
|
|
|
| Sample: |
Cytohesin-3 dimer, 90 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 2 mM MgCl2, 0.1% 2-mercaptoethanol, 5% glycerol, 0.001 mM insitol 1,3,4,5-tetrakis phosphate, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2013 Nov 15
|
Structural Organization and Dynamics of Homodimeric Cytohesin Family Arf GTPase Exchange Factors in Solution and on Membranes.
Structure (2019)
Das S, Malaby AW, Nawrotek A, Zhang W, Zeghouf M, Maslen S, Skehel M, Chakravarthy S, Irving TC, Bilsel O, Cherfils J, Lambright DG
|
| RgGuinier |
5.1 |
nm |
| Dmax |
25.7 |
nm |
| VolumePorod |
168 |
nm3 |
|
|