UniProt ID: Q6NUC7 (2-145) Nucleoplasmin core + A2
UniProt ID: Q6AZJ8 (2-130) Histone H2A (ΔAla127)
UniProt ID: P02281 (5-126) Histone H2B 1.1 (Ser33Thr)
|
|
|

|
| Sample: |
Nucleoplasmin core + A2 pentamer, 81 kDa Xenopus laevis protein
Histone H2A (ΔAla127) pentamer, 69 kDa Xenopus laevis protein
Histone H2B 1.1 (Ser33Thr) pentamer, 67 kDa Xenopus laevis protein
|
| Buffer: |
20 mM Tris. 150 mM NaCl, 1 mM EDTA, 5 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2016 Jan 7
|
Dynamic intramolecular regulation of the histone chaperone nucleoplasmin controls histone binding and release.
Nat Commun 8(1):2215 (2017)
Warren C, Matsui T, Karp JM, Onikubo T, Cahill S, Brenowitz M, Cowburn D, Girvin M, Shechter D
|
| RgGuinier |
4.4 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
402 |
nm3 |
|
|
UniProt ID: None (2-136) Histone H3
UniProt ID: P62799 (2-103) Histone H4
UniProt ID: Q6AZJ8 (2-130) Histone H2a
UniProt ID: Q92130 (5-126) Histone H2b
UniProt ID: None (None-None) 147bp 601 Widom sequence
|
|
|
|
| Sample: |
Histone H3 dimer, 31 kDa Xenopus laevis protein
Histone H4 dimer, 22 kDa Xenopus laevis protein
Histone H2a dimer, 28 kDa Xenopus laevis protein
Histone H2b dimer, 27 kDa Xenopus laevis protein
147bp 601 Widom sequence monomer, 45 kDa synthetic construct DNA
|
| Buffer: |
15 mM HEPES, 200 mM NaCl, pH: 7.3 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Dec 11
|
A Tail-Based Mechanism Drives Nucleosome Demethylation by the LSD2/NPAC Multimeric Complex.
Cell Rep 27(2):387-399.e7 (2019)
Marabelli C, Marrocco B, Pilotto S, Chittori S, Picaud S, Marchese S, Ciossani G, Forneris F, Filippakopoulos P, Schoehn G, Rhodes D, Subramaniam S, Mattevi A
|
| RgGuinier |
4.4 |
nm |
| Dmax |
12.8 |
nm |
| VolumePorod |
353 |
nm3 |
|
|
UniProt ID: Q8NB78 (31-822) Lysyne-specific Demethylase LSD2
UniProt ID: Q49A26 (205-553) NPAC linker+DH (delta-205)
UniProt ID: None (2-136) Histone H3
UniProt ID: P62799 (2-103) Histone H4
UniProt ID: Q6AZJ8 (2-130) Histone H2a
UniProt ID: Q92130 (5-126) Histone H2b
UniProt ID: None (None-None) 147bp 601 Widom sequence
|
|
|
|
| Sample: |
Lysyne-specific Demethylase LSD2 monomer, 89 kDa Homo sapiens protein
NPAC linker+DH (delta-205) tetramer, 150 kDa Homo sapiens protein
Histone H3 dimer, 31 kDa Xenopus laevis protein
Histone H4 dimer, 22 kDa Xenopus laevis protein
Histone H2a dimer, 28 kDa Xenopus laevis protein
Histone H2b dimer, 27 kDa Xenopus laevis protein
147bp 601 Widom sequence monomer, 45 kDa synthetic construct DNA
|
| Buffer: |
15 mM HEPES, 200 mM NaCl, pH: 7.3 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Dec 11
|
A Tail-Based Mechanism Drives Nucleosome Demethylation by the LSD2/NPAC Multimeric Complex.
Cell Rep 27(2):387-399.e7 (2019)
Marabelli C, Marrocco B, Pilotto S, Chittori S, Picaud S, Marchese S, Ciossani G, Forneris F, Filippakopoulos P, Schoehn G, Rhodes D, Subramaniam S, Mattevi A
|
| RgGuinier |
7.8 |
nm |
| Dmax |
30.4 |
nm |
| VolumePorod |
1100 |
nm3 |
|
|
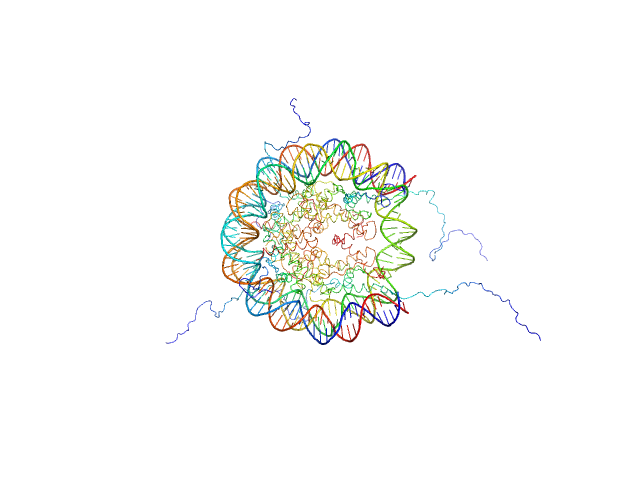
UniProt ID: P84233 (1-135) Histone H3.2
UniProt ID: P62799 (1-103) Histone H4
UniProt ID: Q6AZJ8 (1-130) Histone H2A
UniProt ID: P02281 (1-126) Histone H2B 1.1
UniProt ID: None (None-None) Widom 601 DNA sequence
|
|
|
|
| Sample: |
Histone H3.2 dimer, 31 kDa Xenopus laevis protein
Histone H4 dimer, 23 kDa Xenopus laevis protein
Histone H2A dimer, 28 kDa Xenopus laevis protein
Histone H2B 1.1 dimer, 28 kDa Xenopus laevis protein
Widom 601 DNA sequence dimer, 89 kDa Escherichia coli DNA
|
| Buffer: |
20 mM Tris-HCl , 50 mM KCl, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Dec 2
|
BCL7 proteins, novel subunits of the mammalian SWI/SNF complex, bind the nucleosome core particle
Asgar Abbas kazrani
|
| RgGuinier |
4.3 |
nm |
| Dmax |
12.3 |
nm |
| VolumePorod |
350 |
nm3 |
|
|
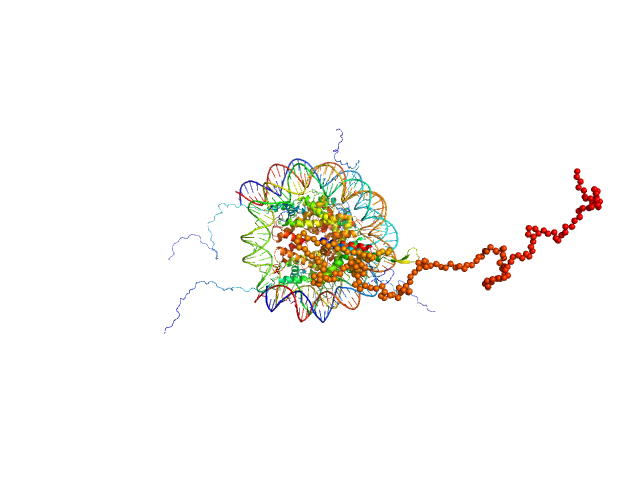
UniProt ID: Q4VC05-2 (1-231) Isoform 2 of B-cell CLL/lymphoma 7 protein family member A
UniProt ID: P84233 (1-135) Histone H3.2
UniProt ID: P62799 (1-103) Histone H4
UniProt ID: Q6AZJ8 (1-130) Histone H2A
UniProt ID: P02281 (1-126) Histone H2B 1.1
UniProt ID: None (None-None) Widom 601 DNA sequence
|
|
|
|
| Sample: |
Isoform 2 of B-cell CLL/lymphoma 7 protein family member A monomer, 25 kDa Homo sapiens protein
Histone H3.2 dimer, 31 kDa Xenopus laevis protein
Histone H4 dimer, 23 kDa Xenopus laevis protein
Histone H2A dimer, 28 kDa Xenopus laevis protein
Histone H2B 1.1 dimer, 28 kDa Xenopus laevis protein
Widom 601 DNA sequence dimer, 89 kDa Escherichia coli DNA
|
| Buffer: |
20 mM Tris-HCl , 50 mM KCl, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Dec 2
|
BCL7 proteins, novel subunits of the mammalian SWI/SNF complex, bind the nucleosome core particle
Asgar Abbas kazrani
|
| RgGuinier |
4.7 |
nm |
| Dmax |
24.4 |
nm |
| VolumePorod |
400 |
nm3 |
|
|