|
|
|
|
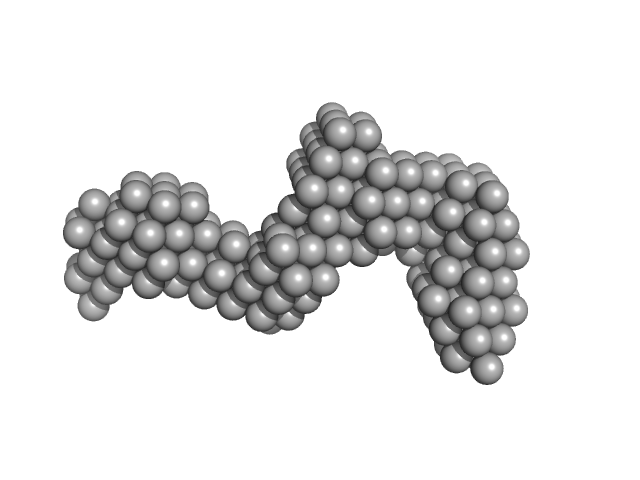
|
| Sample: |
Oxidized tannin macromolecules (DP7, average polymerization 6.3) in water-ethanol solution (water fraction 80%) heptamer, 2 kDa
|
| Buffer: |
water-ethanol solution (water fraction 80%), pH: 7 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2007 Oct 12
|
Rigidity, conformation, and solvation of native and oxidized tannin macromolecules in water-ethanol solution.
J Chem Phys 130(24):245103 (2009)
Zanchi D, Konarev PV, Tribet C, Baron A, Svergun DI, Guyot S
|
| RgGuinier |
1.2 |
nm |
| Dmax |
3.6 |
nm |
|
|
|
|
|
|
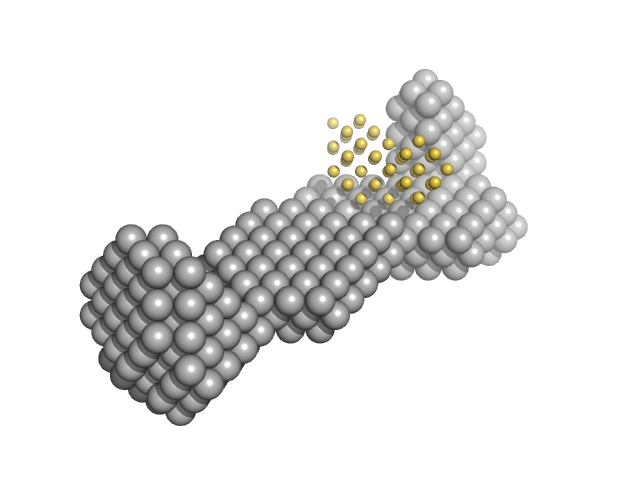
|
| Sample: |
Replication origin-binding protein dimer, 81 kDa Human herpesvirus 1 … protein
15-bp DNA monomer, 9 kDa DNA
|
| Buffer: |
20 mM Tris-HCl, 50 mM NaCl, 5 mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 May 27
|
Structural and biophysical characterization of the proteins interacting with the herpes simplex virus 1 origin of replication.
J Biol Chem 284(24):16343-16353 (2009)
Manolaridis I, Mumtsidu E, Konarev P, Makhov AM, Fullerton SW, Sinz A, Kalkhof S, McGeehan JE, Cary PD, Griffith JD, Svergun D, Kneale GG, Tucker PA
|
| RgGuinier |
3.6 |
nm |
| Dmax |
15.0 |
nm |
|
|
|
|
|
|

|
| Sample: |
SRB2m dimer, 33 kDa RNA
|
| Buffer: |
Hepes, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2007 Oct 24
|
Characterization of a fluorophore binding RNA aptamer by fluorescence correlation spectroscopy and small angle X-ray scattering.
Anal Biochem 389(1):52-62 (2009)
Werner A, Konarev PV, Svergun DI, Hahn U
|
| RgGuinier |
2.8 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
38 |
nm3 |
|
|
|
|
|
|
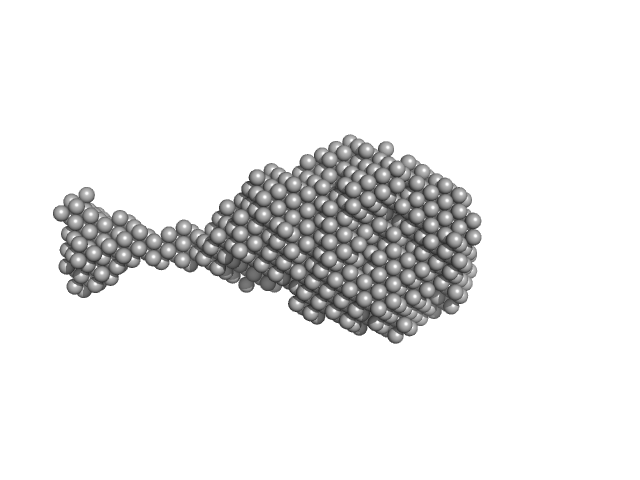
|
| Sample: |
SRB2m dimer, 33 kDa RNA
|
| Buffer: |
Hepes, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2007 Oct 24
|
Characterization of a fluorophore binding RNA aptamer by fluorescence correlation spectroscopy and small angle X-ray scattering.
Anal Biochem 389(1):52-62 (2009)
Werner A, Konarev PV, Svergun DI, Hahn U
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
31 |
nm3 |
|
|
|
|
|
|
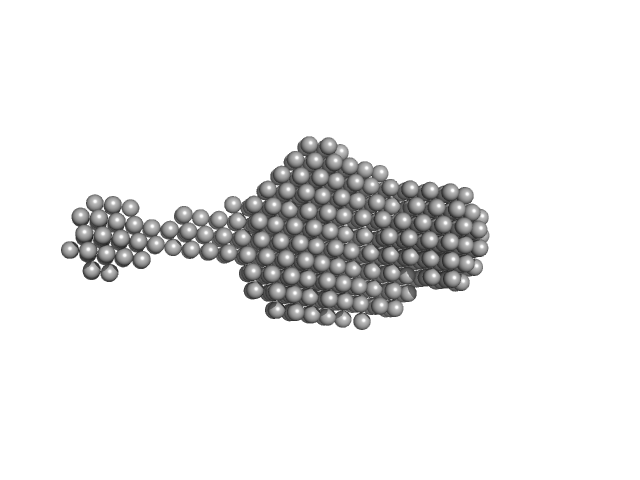
|
| Sample: |
SRB2m dimer, 33 kDa RNA
|
| Buffer: |
Hepes, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2007 Oct 24
|
Characterization of a fluorophore binding RNA aptamer by fluorescence correlation spectroscopy and small angle X-ray scattering.
Anal Biochem 389(1):52-62 (2009)
Werner A, Konarev PV, Svergun DI, Hahn U
|
| RgGuinier |
2.3 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
24 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
ProNGF dimer, 45 kDa Mouse submandibulary glands protein
|
| Buffer: |
50 mM Naphosphat 0.5 M Ammonium Sulfate(NH4)2SO4, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2007 Oct 25
|
Intrinsic structural disorder of mouse proNGF.
Proteins 75(4):990-1009 (2009)
Paoletti F, Covaceuszach S, Konarev PV, Gonfloni S, Malerba F, Schwarz E, Svergun DI, Cattaneo A, Lamba D
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
92 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Endoglin tetramer, 243 kDa Homo sapiens protein
|
| Buffer: |
10 mM Tris–HCl, 50 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2008 Oct 17
|
Structural and functional characterization of soluble endoglin receptor
Biochemical and Biophysical Research Communications 383(4):386-391 (2009)
Le B, Franke D, Svergun D, Han T, Hwang H, Kim K
|
| RgGuinier |
7.6 |
nm |
| Dmax |
26.0 |
nm |
| VolumePorod |
434 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Endoglin dimer, 121 kDa Homo sapiens protein
|
| Buffer: |
10 mM Tris–HCl, 50 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2008 Oct 17
|
Structural and functional characterization of soluble endoglin receptor
Biochemical and Biophysical Research Communications 383(4):386-391 (2009)
Le B, Franke D, Svergun D, Han T, Hwang H, Kim K
|
| RgGuinier |
4.7 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
173 |
nm3 |
|
|
|
|
|
|
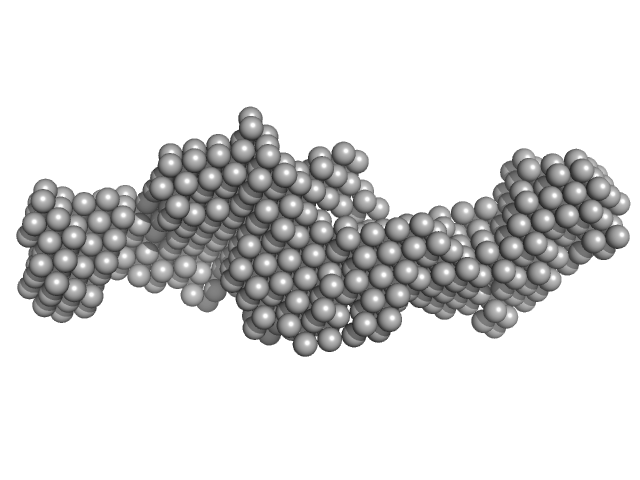
|
| Sample: |
Nuclear fragile X mental retardation-interacting protein 2 monomer, 59 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl pH 8.0, 5 mM TCEP, pH: |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2007 Oct 11
|
A study of the ultrastructure of fragile-X-related proteins.
Biochem J 419(2):347-57 (2009)
Sjekloća L, Konarev PV, Eccleston J, Taylor IA, Svergun DI, Pastore A
|
| RgGuinier |
4.1 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
92 |
nm3 |
|
|
|
|
|
|
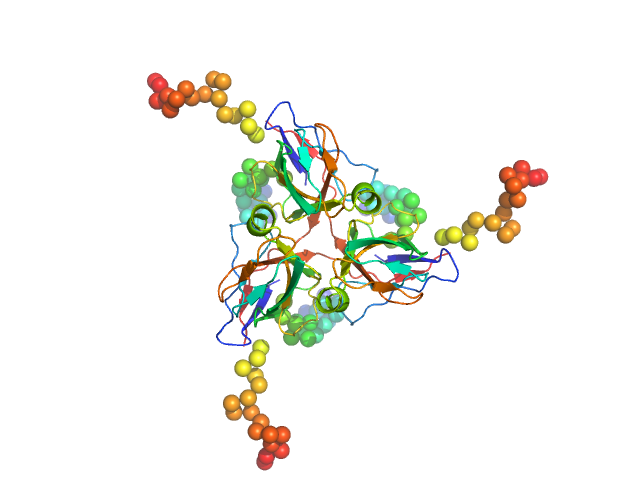
|
| Sample: |
Deoxyuridine 5'-triphosphate nucleotidohydrolase, mitochondrial trimer, 80 kDa Homo sapiens protein
|
| Buffer: |
20 mM citrate buffer, 1 mM DTT, 0.1 mM PMSF and 1 mM MgCl2, pH: 5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2005 Feb 1
|
Molecular shape and prominent role of β-strand swapping in organization of dUTPase oligomers
FEBS Letters 583(5):865-871 (2009)
Takács E, Barabás O, Petoukhov M, Svergun D, Vértessy B
|
| RgGuinier |
2.6 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
101 |
nm3 |
|
|