|
|
|
|
|
| Sample: |
DHPC - 1,2-dihexanoyl-sn-glycero-3-phosphocholine None, lipid
POPC - 2-oleoyl-1-palmitoyl-sn-glyecro-3-phosphocholine None, lipid
POPG - 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) None, lipid
|
| Buffer: |
Tris buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Mar 17
|
Expanding the Toolbox for Bicelle-Forming Surfactant–Lipid Mixtures
Molecules 27(21):7628 (2022)
Giudice R, Paracini N, Laursen T, Blanchet C, Roosen-Runge F, Cárdenas M
|
|
|
|
|
|
|
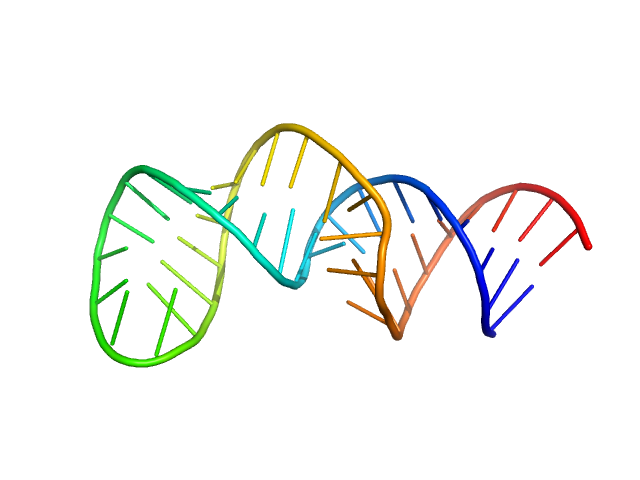
|
| Sample: |
The second exon splicing silencer 2p monomer, 14 kDa RNA
|
| Buffer: |
20 mM Bis-Tris, 20 mM NaCl, pH: 6.2 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2017 Nov 1
|
Encoded Conformational Dynamics of the HIV Splice Site A3 Regulatory Locus: Implications for Differential Binding of hnRNP Splicing Auxiliary Factors.
J Mol Biol 434(18):167728 (2022)
Chiu LY, Emery A, Jain N, Sugarman A, Kendrick N, Luo L, Ford W, Swanstrom R, Tolbert BS
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.9 |
nm |
| VolumePorod |
21 |
nm3 |
|
|
|
|
|
|
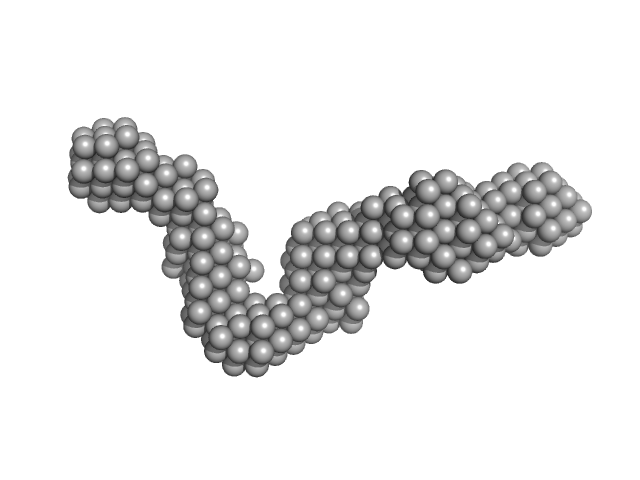
|
| Sample: |
40-mer single stranded inhibitory DNA monomer, 12 kDa DNA
|
| Buffer: |
50 mM phosphate pH 6.0, 200 mM NaCl, 2 mM β-mercaptoethanol (β-ME), 5% glycerol, 200 µM Na2-EDTA, pH: 6 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Aug 8
|
Small-Angle X-ray Scattering (SAXS) Measurements of APOBEC3G Provide Structural Basis for Binding of Single-Stranded DNA and Processivity
Viruses 14(9):1974 (2022)
Barzak F, Ryan T, Mohammadzadeh N, Harjes S, Kvach M, Kurup H, Krause K, Chelico L, Filichev V, Harjes E, Jameson G
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.8 |
nm |
| VolumePorod |
26 |
nm3 |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDNU4_fit1_model1.png)
|
| Sample: |
Di[3-deoxy-D-manno-octulosonyl]-lipid A (ammonium salt), unidentified lipid
|
| Buffer: |
water, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2018 Apr 1
|
Antimicrobial Peptide Mechanism Studied by Scattering-Guided Molecular Dynamics Simulation.
J Phys Chem B (2022)
Allsopp R, Pavlova A, Cline T, Salyapongse AM, Gillilan RE, Di YP, Deslouches B, Klauda JB, Gumbart JC, Tristram-Nagle S
|
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDNV4_fit1_model1.png)
|
| Sample: |
Di[3-deoxy-D-manno-octulosonyl]-lipid A (ammonium salt) plus WLBU2, unidentified lipid
|
| Buffer: |
water, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2018 Apr 1
|
Antimicrobial Peptide Mechanism Studied by Scattering-Guided Molecular Dynamics Simulation.
J Phys Chem B (2022)
Allsopp R, Pavlova A, Cline T, Salyapongse AM, Gillilan RE, Di YP, Deslouches B, Klauda JB, Gumbart JC, Tristram-Nagle S
|
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDNW4_fit1_model1.png)
|
| Sample: |
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine, 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(10-rac-glycerol) sodium salt, 10,30-bis-[1,2-dioleoyl-sn-glycero-3-phospho]-sn-glycerol (7:2:1), lipid
|
| Buffer: |
water, pH: 7 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2021 Jun 23
|
Antimicrobial Peptide Mechanism Studied by Scattering-Guided Molecular Dynamics Simulation.
J Phys Chem B (2022)
Allsopp R, Pavlova A, Cline T, Salyapongse AM, Gillilan RE, Di YP, Deslouches B, Klauda JB, Gumbart JC, Tristram-Nagle S
|
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDNX4_fit1_model1.png)
|
| Sample: |
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanol, 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(10-rac-glycerol) sodium salt, 10,30-bis-[1,2-dioleoyl-sn-glycero-3-phospho]-sn-glycerol sodium salt, WLBU2, synthetic construct lipid
|
| Buffer: |
water, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2018 Apr 1
|
Antimicrobial Peptide Mechanism Studied by Scattering-Guided Molecular Dynamics Simulation.
J Phys Chem B (2022)
Allsopp R, Pavlova A, Cline T, Salyapongse AM, Gillilan RE, Di YP, Deslouches B, Klauda JB, Gumbart JC, Tristram-Nagle S
|
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDNY4_fit1_model1.png)
|
| Sample: |
1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(10-rac-glycerol), 1,2-dioleoyl-3-trimethylammonium-propane, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine, 10,30-bis-[1,2-dioleoyl-sn-glycero-3-phos, lipid
|
| Buffer: |
water, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2018 Apr 1
|
Antimicrobial Peptide Mechanism Studied by Scattering-Guided Molecular Dynamics Simulation.
J Phys Chem B (2022)
Allsopp R, Pavlova A, Cline T, Salyapongse AM, Gillilan RE, Di YP, Deslouches B, Klauda JB, Gumbart JC, Tristram-Nagle S
|
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDNZ4_fit1_model1.png)
|
| Sample: |
1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(10-rac-glycerol), 1,2-dioleoyl-3-trimethylammonium-propane, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine, 10,30-bis-[1,2-dioleoyl-sn-glycero-3-phos, synthetic construct lipid
|
| Buffer: |
water, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2018 Apr 1
|
Antimicrobial Peptide Mechanism Studied by Scattering-Guided Molecular Dynamics Simulation.
J Phys Chem B (2022)
Allsopp R, Pavlova A, Cline T, Salyapongse AM, Gillilan RE, Di YP, Deslouches B, Klauda JB, Gumbart JC, Tristram-Nagle S
|
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDN25_fit1_model1.png)
|
| Sample: |
Lipopolysaccharide, Pseudomonas aeruginosa lipid
|
| Buffer: |
water, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2018 Jun 1
|
Antimicrobial Peptide Mechanism Studied by Scattering-Guided Molecular Dynamics Simulation.
J Phys Chem B (2022)
Allsopp R, Pavlova A, Cline T, Salyapongse AM, Gillilan RE, Di YP, Deslouches B, Klauda JB, Gumbart JC, Tristram-Nagle S
|
|
|