|
|
|
|
|
| Sample: |
Sigma-Aldrich polystyrene nanospheres with a diameter of 100 nm monomer, 0 kDa synthetic construct
|
| Buffer: |
water/detergent solution (0.0125 % (v/v) NovaChem), pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Jun 19
|
AF4-to-SAXS: expanded characterization of nanoparticles and proteins at the P12 BioSAXS beamline.
J Synchrotron Radiat (2025)
Da Vela S, Bartels K, Franke D, Soloviov D, Gräwert T, Molodenskiy D, Kolb B, Wilhelmy C, Drexel R, Meier F, Haas H, Langguth P, Graewert MA
|
| RgGuinier |
47.7 |
nm |
| Dmax |
111.0 |
nm |
|
|
|
|
|
|
|
| Sample: |
Thermo Fisher polystyrene nanospheres with a diameter of 20 nm monomer, 1 kDa synthetic construct
|
| Buffer: |
water/detergent solution (0.0125 % (v/v) NovaChem), pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Jun 19
|
AF4-to-SAXS: expanded characterization of nanoparticles and proteins at the P12 BioSAXS beamline.
J Synchrotron Radiat (2025)
Da Vela S, Bartels K, Franke D, Soloviov D, Gräwert T, Molodenskiy D, Kolb B, Wilhelmy C, Drexel R, Meier F, Haas H, Langguth P, Graewert MA
|
| RgGuinier |
9.0 |
nm |
| Dmax |
25.0 |
nm |
|
|
|
|
|
|
|
| Sample: |
I-RNA, Top-strand monomer, 7 kDa RNA
I-RNA, Bottom-strand monomer, 7 kDa RNA
|
| Buffer: |
25 mM sodium phosphate 25 mM sodium chloride, pH: 6.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Nov 9
|
Unique conformational dynamics and protein recognition of A-to-I hyper-edited dsRNA.
Nucleic Acids Res 53(12) (2025)
Müller-Hermes C, Piomponi V, Hilber S, Asami S, Kreutz C, Bussi G, Sattler M
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.9 |
nm |
| VolumePorod |
17 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
A-RNA, Top-Strand monomer, 6 kDa RNA
A-RNA, Bottom-Strand monomer, 6 kDa RNA
|
| Buffer: |
25 mM sodium phosphate 25 mM sodium chloride, pH: 6.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Nov 9
|
Unique conformational dynamics and protein recognition of A-to-I hyper-edited dsRNA.
Nucleic Acids Res 53(12) (2025)
Müller-Hermes C, Piomponi V, Hilber S, Asami S, Kreutz C, Bussi G, Sattler M
|
| RgGuinier |
1.8 |
nm |
| Dmax |
6.2 |
nm |
| VolumePorod |
17 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Single stranded 9-mer RNA: tristetraprolin (TTP) optimal target sequence monomer, 3 kDa synthetic construct RNA
|
| Buffer: |
20 mM sodium phosphate, 100 mM KCl, 25 µM ZnSO4, 2 mM β-mercaptoethanol, 5 % glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Mar 19
|
Solution scattering of the tandem zinc finger (TZF) domain from the tristetraprolin (TTP) family member found in Selaginella moellendorffii (spikemoss) in the presence and absence of 9-mer RNA
Stephanie Hicks
|
| RgGuinier |
1.0 |
nm |
| Dmax |
3.5 |
nm |
| VolumePorod |
5 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Single stranded 9-mer RNA: tristetraprolin (TTP) optimal target sequence monomer, 3 kDa synthetic construct RNA
|
| Buffer: |
20 mM sodium phosphate, 100 mM KCl, 25 µM ZnSO4, 2 mM β-mercaptoethanol, 5 % glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Mar 19
|
Solution scattering of the tandem zinc finger (TZF) domain from the tristetraprolin (TTP) family member found in Selaginella moellendorffii (spikemoss) in the presence and absence of 9-mer RNA
Stephanie Hicks
|
| RgGuinier |
1.0 |
nm |
| Dmax |
3.5 |
nm |
| VolumePorod |
5 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Single stranded 9-mer RNA: tristetraprolin (TTP) optimal target sequence monomer, 3 kDa synthetic construct RNA
|
| Buffer: |
20 mM sodium phosphate, 100 mM KCl, 25 µM ZnSO4, 2 mM β-mercaptoethanol, 5 % glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Mar 19
|
Solution scattering of the tandem zinc finger (TZF) domain from the tristetraprolin (TTP) family member found in Selaginella moellendorffii (spikemoss) in the presence and absence of 9-mer RNA
Stephanie Hicks
|
| RgGuinier |
1.0 |
nm |
| Dmax |
3.2 |
nm |
| VolumePorod |
5 |
nm3 |
|
|
|
|
|
|
|
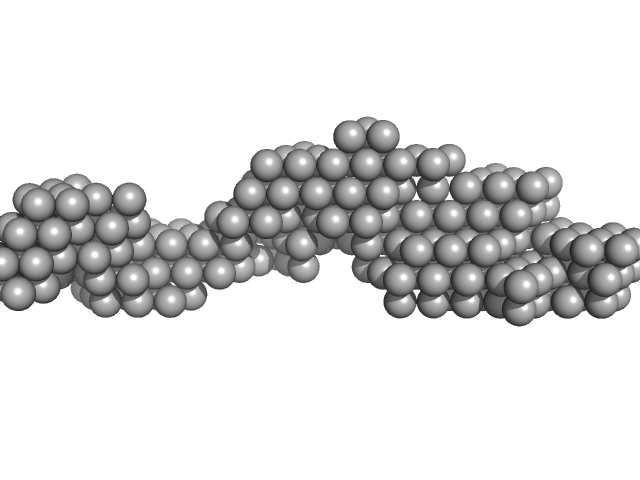
| Sample: |
BC120 RNA monomer monomer, 39 kDa RNA
|
| Buffer: |
phosphate buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Sep 22
|
Alu RNA pseudoknot alterations influence SRP9/SRP14 association.
RNA (2025)
Gussakovsky D, Brown MJF, Pereira HS, Meier M, Padilla-Meier GP, Black NA, Booy EP, Stetefeld J, Patel TR, McKenna SA
|
| RgGuinier |
4.0 |
nm |
| Dmax |
14.3 |
nm |
| VolumePorod |
57 |
nm3 |
|
|
|
|
|
|

|
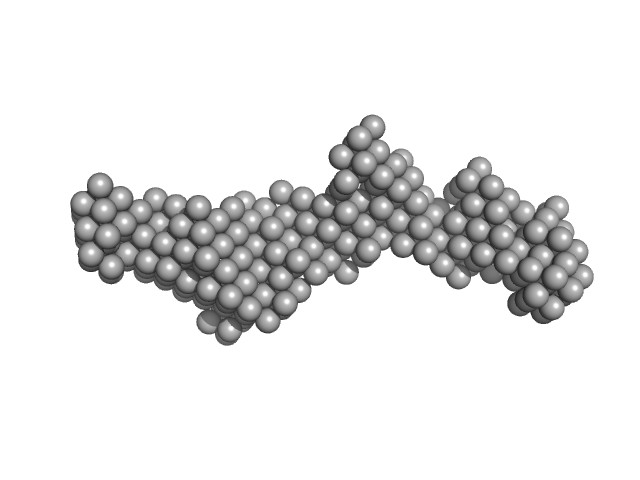
| Sample: |
EB120 RNA monomer monomer, 39 kDa RNA
|
| Buffer: |
phosphate buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Sep 22
|
Alu RNA pseudoknot alterations influence SRP9/SRP14 association.
RNA (2025)
Gussakovsky D, Brown MJF, Pereira HS, Meier M, Padilla-Meier GP, Black NA, Booy EP, Stetefeld J, Patel TR, McKenna SA
|
| RgGuinier |
4.2 |
nm |
| Dmax |
13.6 |
nm |
| VolumePorod |
79 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
BC120 G25C RNA monomer, 39 kDa Homo sapiens RNA
|
| Buffer: |
phosphate buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Sep 22
|
Alu RNA pseudoknot alterations influence SRP9/SRP14 association.
RNA (2025)
Gussakovsky D, Brown MJF, Pereira HS, Meier M, Padilla-Meier GP, Black NA, Booy EP, Stetefeld J, Patel TR, McKenna SA
|
| RgGuinier |
4.1 |
nm |
| Dmax |
13.8 |
nm |
| VolumePorod |
63 |
nm3 |
|
|