|
|
|
|
|
| Sample: |
25 base-paired RNA double helix monomer, 16 kDa RNA
|
| Buffer: |
400 mM KCl, 20 mM KMOPS, 20 µM EDTA, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2017 May 13
|
Salt Dependence of A-Form RNA Duplexes: Structures and Implications.
J Phys Chem B 123(46):9773-9785 (2019)
Chen YL, Pollack L
|
|
|
|
|
|
|
|
| Sample: |
25 base-paired DNA double helix monomer, 15 kDa DNA
|
| Buffer: |
400 mM KCl, 20 mM KMOPS, 20 µM EDTA, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2017 Apr 16
|
Salt Dependence of A-Form RNA Duplexes: Structures and Implications.
J Phys Chem B 123(46):9773-9785 (2019)
Chen YL, Pollack L
|
|
|
|
|
|
|
|
| Sample: |
25 base-paired DNA double helix monomer, 15 kDa DNA
|
| Buffer: |
0.5 mM MgCl2, 20 mM KMOPS, 20 µM EDTA, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2017 Apr 16
|
Salt Dependence of A-Form RNA Duplexes: Structures and Implications.
J Phys Chem B 123(46):9773-9785 (2019)
Chen YL, Pollack L
|
|
|
|
|
|
|
|
| Sample: |
25 base-paired DNA double helix monomer, 15 kDa DNA
|
| Buffer: |
2.0 mM MgCl2, 20 mM KMOPS, 20 µM EDTA, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2017 Apr 16
|
Salt Dependence of A-Form RNA Duplexes: Structures and Implications.
J Phys Chem B 123(46):9773-9785 (2019)
Chen YL, Pollack L
|
|
|
|
|
|
|

|
| Sample: |
Xrn1 resistance RNA2 from Zika virus monomer, 22 kDa Zika virus RNA
|
| Buffer: |
20mM Tris-HCl, 100mM NaCl, 5mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2016 Dec 9
|
Long non-coding subgenomic flavivirus RNAs have extended 3D structures and are flexible in solution.
EMBO Rep 20(11):e47016 (2019)
Zhang Y, Zhang Y, Liu ZY, Cheng ML, Ma J, Wang Y, Qin CF, Fang X
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
34 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Xrn1 resistance RNA1 from Dengue virus 2 monomer, 21 kDa Dengue virus 2 RNA
|
| Buffer: |
20mM Tris-HCl, 100mM NaCl, 5mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at 14-ID-B (BioCARS), Advanced Photon Source (APS), Argonne National Laboratory on 2016 Dec 9
|
Long non-coding subgenomic flavivirus RNAs have extended 3D structures and are flexible in solution.
EMBO Rep 20(11):e47016 (2019)
Zhang Y, Zhang Y, Liu ZY, Cheng ML, Ma J, Wang Y, Qin CF, Fang X
|
| RgGuinier |
2.2 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
31 |
nm3 |
|
|
|
|
|
|
|
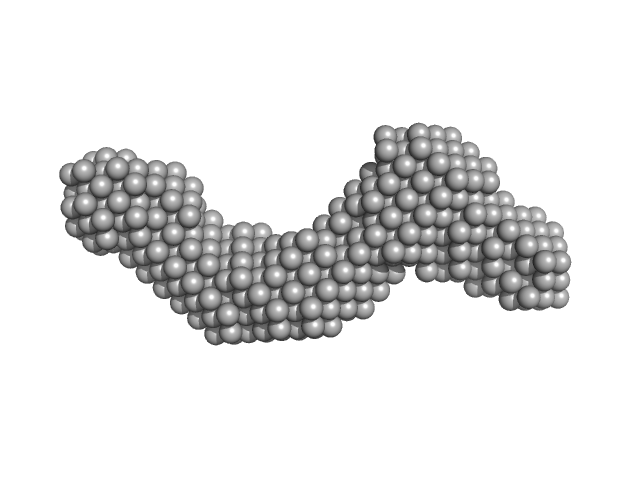
| Sample: |
Xrn1 resistance RNA1-2 from Dengue virus 2 monomer, 46 kDa Dengue virus 2 RNA
|
| Buffer: |
20mM Tris-HCl, 100mM NaCl, 5mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2017 Apr 2
|
Long non-coding subgenomic flavivirus RNAs have extended 3D structures and are flexible in solution.
EMBO Rep 20(11):e47016 (2019)
Zhang Y, Zhang Y, Liu ZY, Cheng ML, Ma J, Wang Y, Qin CF, Fang X
|
| RgGuinier |
3.9 |
nm |
| Dmax |
13.4 |
nm |
| VolumePorod |
67 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Xrn1 resistance RNA-1 from West Nile virus monomer, 25 kDa West Nile virus RNA
|
| Buffer: |
20mM Tris-HCl, 100mM NaCl, 5mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2016 Dec 9
|
Long non-coding subgenomic flavivirus RNAs have extended 3D structures and are flexible in solution.
EMBO Rep 20(11):e47016 (2019)
Zhang Y, Zhang Y, Liu ZY, Cheng ML, Ma J, Wang Y, Qin CF, Fang X
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.4 |
nm |
| VolumePorod |
34 |
nm3 |
|
|
|
|
|
|

|
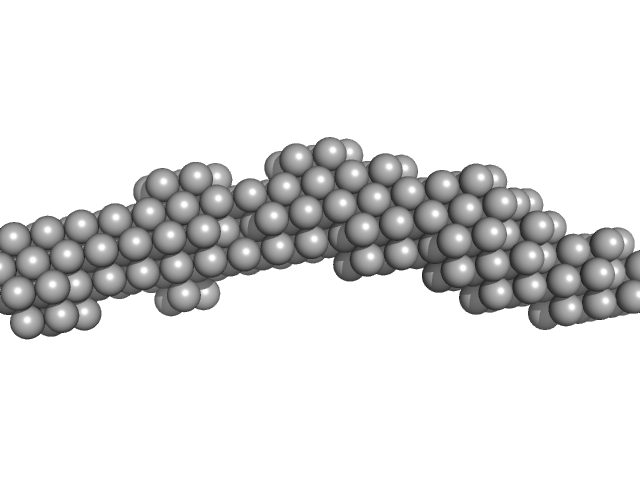
| Sample: |
Xrn1 resistance RNA-1 from Murray Valley Encephalitis monomer, 26 kDa Murray Valley Encephalitis RNA
|
| Buffer: |
20mM Tris-HCl, 100mM NaCl, 5mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2016 Dec 9
|
Long non-coding subgenomic flavivirus RNAs have extended 3D structures and are flexible in solution.
EMBO Rep 20(11):e47016 (2019)
Zhang Y, Zhang Y, Liu ZY, Cheng ML, Ma J, Wang Y, Qin CF, Fang X
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.8 |
nm |
| VolumePorod |
31 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
3'SL from Dengue virus 2 monomer, 31 kDa Dengue virus 2 RNA
|
| Buffer: |
20mM Tris-HCl, 100mM NaCl, 5mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2017 Feb 24
|
Long non-coding subgenomic flavivirus RNAs have extended 3D structures and are flexible in solution.
EMBO Rep 20(11):e47016 (2019)
Zhang Y, Zhang Y, Liu ZY, Cheng ML, Ma J, Wang Y, Qin CF, Fang X
|
| RgGuinier |
3.6 |
nm |
| Dmax |
14.1 |
nm |
| VolumePorod |
40 |
nm3 |
|
|