|
|
|
|
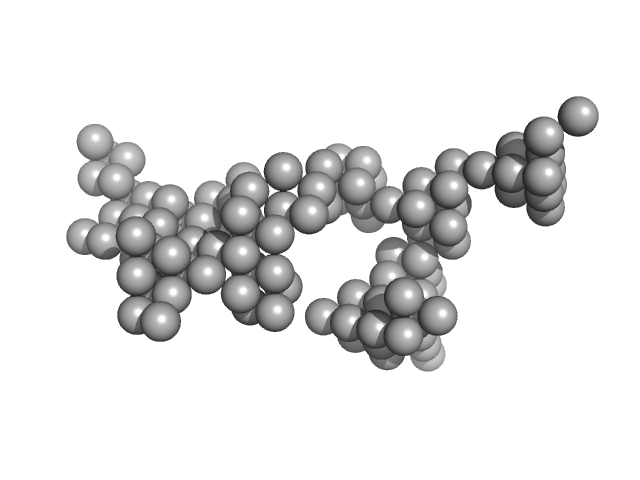
|
| Sample: |
Hydrolytically Degradable Polymer Micelles for Drug Delivery (structure of hydrophobic substituent - Lev-Chol) with Dox (10%) monomer, 125 kDa
|
| Buffer: |
phosphate buffer saline (PBS) (pH 5.0), pH: 5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2012 Aug 19
|
Hydrolytically degradable polymer micelles for drug delivery: a SAXS/SANS kinetic study.
Biomacromolecules 14(11):4061-70 (2013)
Filippov SK, Franklin JM, Konarev PV, Chytil P, Etrych T, Bogomolova A, Dyakonova M, Papadakis CM, Radulescu A, Ulbrich K, Stepanek P, Svergun DI
|
| RgGuinier |
6.9 |
nm |
| Dmax |
23.8 |
nm |
|
|
|
|
|
|
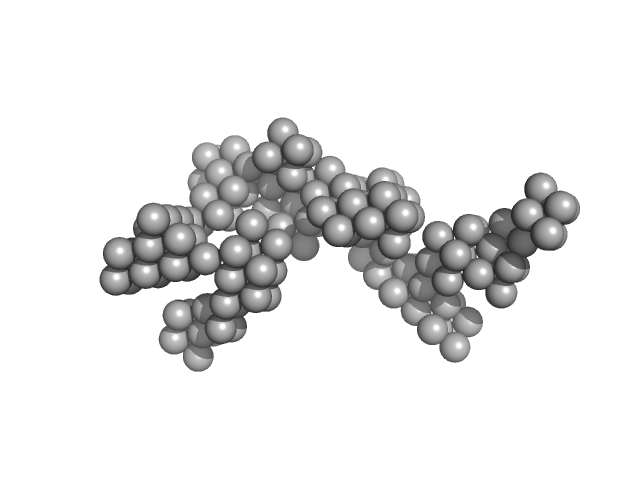
|
| Sample: |
Hydrolytically Degradable Polymer Micelles for Drug Delivery (structure of hydrophobic substituent - cholest-4-en-3-one) without Dox monomer, 125 kDa
|
| Buffer: |
phosphate buffer saline (PBS) (pH 5.0), pH: 5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2012 Aug 16
|
Hydrolytically degradable polymer micelles for drug delivery: a SAXS/SANS kinetic study.
Biomacromolecules 14(11):4061-70 (2013)
Filippov SK, Franklin JM, Konarev PV, Chytil P, Etrych T, Bogomolova A, Dyakonova M, Papadakis CM, Radulescu A, Ulbrich K, Stepanek P, Svergun DI
|
| RgGuinier |
5.9 |
nm |
| Dmax |
19.7 |
nm |
|
|
|
|
|
|
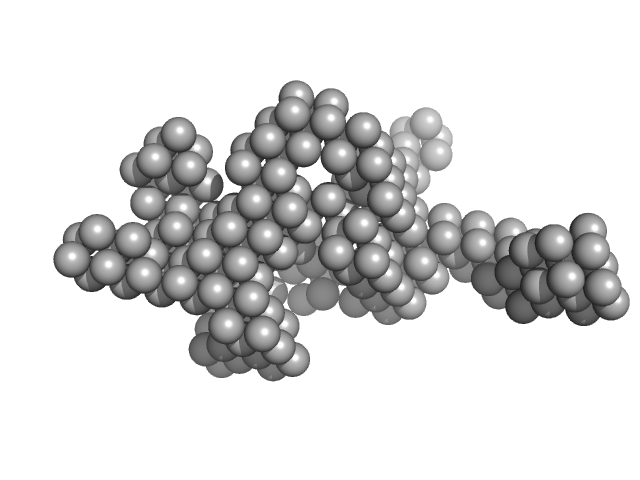
|
| Sample: |
Hydrolytically Degradable Polymer Micelles for Drug Delivery (structure of hydrophobic substituent - cholest-4-en-3-one) with Dox (10%) monomer, 125 kDa
|
| Buffer: |
phosphate buffer saline (PBS) (pH 5.0), pH: 5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2012 Aug 19
|
Hydrolytically degradable polymer micelles for drug delivery: a SAXS/SANS kinetic study.
Biomacromolecules 14(11):4061-70 (2013)
Filippov SK, Franklin JM, Konarev PV, Chytil P, Etrych T, Bogomolova A, Dyakonova M, Papadakis CM, Radulescu A, Ulbrich K, Stepanek P, Svergun DI
|
| RgGuinier |
5.5 |
nm |
| Dmax |
18.3 |
nm |
|
|
|
|
|
|

|
| Sample: |
N-(2-hydroxypropyl)- 31 methacrylamide (HPMA) copolymers with cholesterol 1.4% 0, 16272 kDa
|
| Buffer: |
phosphate buffer saline (PBS) (pH 5.0), pH: 5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Mar 27
|
Macromolecular HPMA-based nanoparticles with cholesterol for solid-tumor targeting: detailed study of the inner structure of a highly efficient drug delivery system.
Biomacromolecules 13(8):2594-604 (2012)
Filippov SK, Chytil P, Konarev PV, Dyakonova M, Papadakis C, Zhigunov A, Plestil J, Stepanek P, Etrych T, Ulbrich K, Svergun DI
|
| RgGuinier |
6.2 |
nm |
| Dmax |
22.0 |
nm |
|
|
|
|
|
|
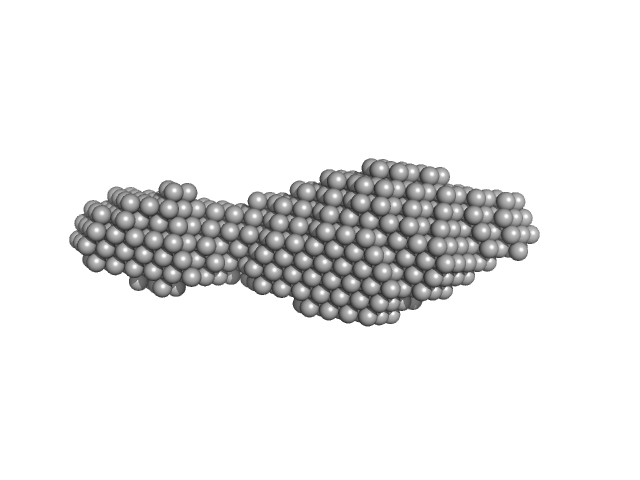
|
| Sample: |
N-(2-hydroxypropyl)- 31 methacrylamide (HPMA) copolymers with Cholesterol (2.7%) 0, 16740 kDa
|
| Buffer: |
phosphate buffer saline (PBS) (pH 7.2), pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Mar 27
|
Macromolecular HPMA-based nanoparticles with cholesterol for solid-tumor targeting: detailed study of the inner structure of a highly efficient drug delivery system.
Biomacromolecules 13(8):2594-604 (2012)
Filippov SK, Chytil P, Konarev PV, Dyakonova M, Papadakis C, Zhigunov A, Plestil J, Stepanek P, Etrych T, Ulbrich K, Svergun DI
|
| RgGuinier |
5.2 |
nm |
| Dmax |
28.1 |
nm |
|
|
|
|
|
|
|
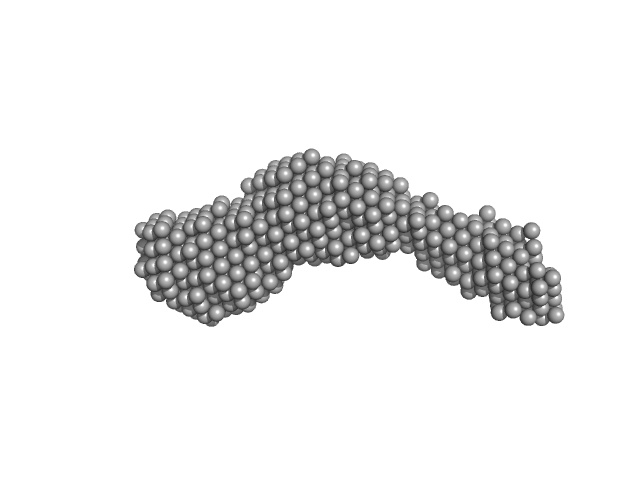
| Sample: |
N-(2-hydroxypropyl)- 31 methacrylamide (HPMA) copolymers with Cholesterol (3.0%) 0, 29520 kDa
|
| Buffer: |
phosphate buffer saline (PBS) (pH 5.0), pH: 5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Mar 27
|
Macromolecular HPMA-based nanoparticles with cholesterol for solid-tumor targeting: detailed study of the inner structure of a highly efficient drug delivery system.
Biomacromolecules 13(8):2594-604 (2012)
Filippov SK, Chytil P, Konarev PV, Dyakonova M, Papadakis C, Zhigunov A, Plestil J, Stepanek P, Etrych T, Ulbrich K, Svergun DI
|
| RgGuinier |
9.4 |
nm |
| Dmax |
43.2 |
nm |
|
|
|
|
|
|
|
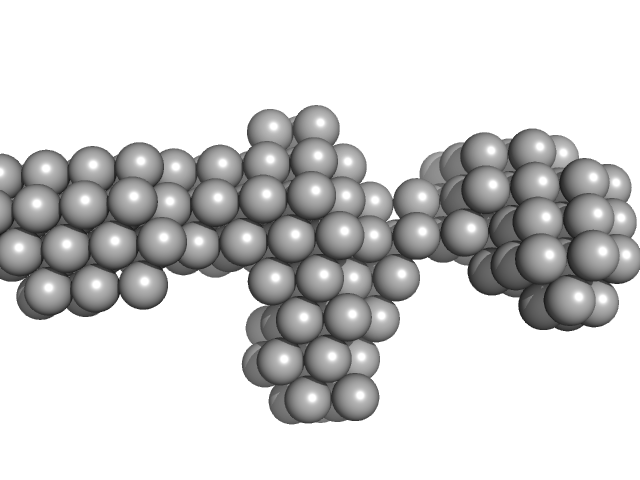
| Sample: |
Native tannin macromolecules (DP7, average polymerization 6.3) in water-ethanol solution (water fraction 0%) heptamer, 2 kDa
|
| Buffer: |
water-ethanol solution (water fraction 0%), pH: 7 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2007 Nov 24
|
Rigidity, conformation, and solvation of native and oxidized tannin macromolecules in water-ethanol solution.
J Chem Phys 130(24):245103 (2009)
Zanchi D, Konarev PV, Tribet C, Baron A, Svergun DI, Guyot S
|
| RgGuinier |
1.0 |
nm |
| Dmax |
3.5 |
nm |
|
|
|
|
|
|

|
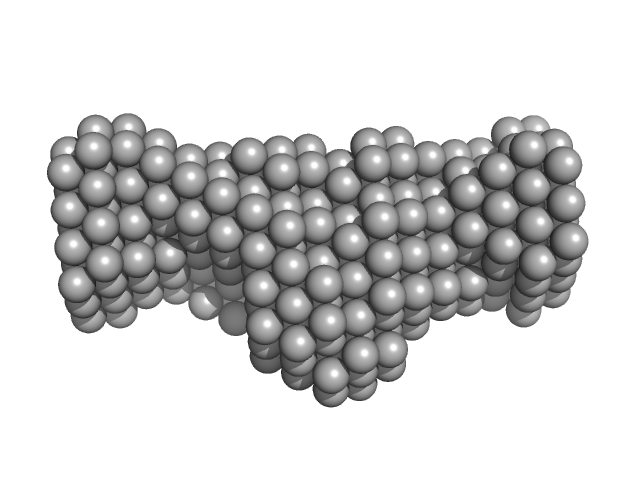
| Sample: |
Native tannin macromolecules (DP7, average polymerization 6.3) in water-ethanol solution (water fraction 60%) heptamer, 2 kDa
|
| Buffer: |
water-ethanol solution (water fraction 60%), pH: 7 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2007 Nov 24
|
Rigidity, conformation, and solvation of native and oxidized tannin macromolecules in water-ethanol solution.
J Chem Phys 130(24):245103 (2009)
Zanchi D, Konarev PV, Tribet C, Baron A, Svergun DI, Guyot S
|
| RgGuinier |
1.0 |
nm |
| Dmax |
3.2 |
nm |
|
|
|
|
|
|
|
| Sample: |
Native tannin macromolecules (DP7, average polymerization 6.3) in water-ethanol solution (water fraction 100%) heptamer, 2 kDa
|
| Buffer: |
water-ethanol solution (water fraction 100%), pH: 7 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2007 Nov 24
|
Rigidity, conformation, and solvation of native and oxidized tannin macromolecules in water-ethanol solution.
J Chem Phys 130(24):245103 (2009)
Zanchi D, Konarev PV, Tribet C, Baron A, Svergun DI, Guyot S
|
| RgGuinier |
1.0 |
nm |
| Dmax |
3.1 |
nm |
|
|
|
|
|
|

|
| Sample: |
Oxidized tannin macromolecules (DP7, average polymerization 6.3) in water-ethanol solution (water fraction 100%) heptamer, 2 kDa
|
| Buffer: |
water-ethanol solution (water fraction 100%), pH: 7 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2007 Oct 12
|
Rigidity, conformation, and solvation of native and oxidized tannin macromolecules in water-ethanol solution.
J Chem Phys 130(24):245103 (2009)
Zanchi D, Konarev PV, Tribet C, Baron A, Svergun DI, Guyot S
|
| RgGuinier |
1.0 |
nm |
| Dmax |
3.3 |
nm |
|
|