|
|
|
|
|
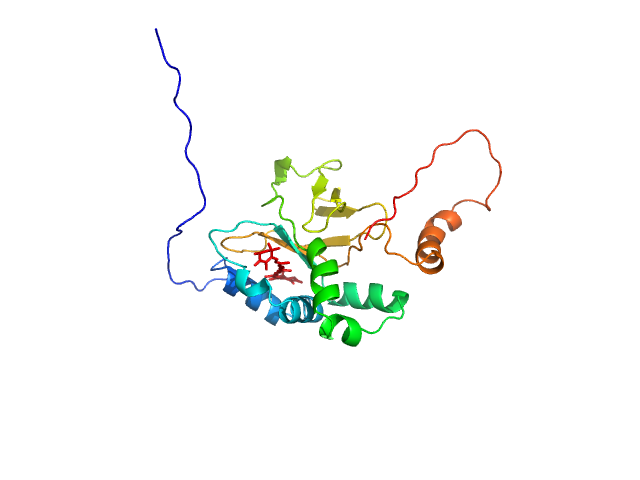
| Sample: |
50S ribosomal protein L11 monomer, 16 kDa Thermus thermophilus protein
|
| Buffer: |
10 mM sodium cacodylate, 100 mM KCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2016 Dec 9
|
Chaotic advection mixer for capturing transient states of diverse biological macromolecular systems with time-resolved small-angle X-ray scattering
IUCrJ 10(3):363-375 (2023)
Zielinski K, Katz A, Calvey G, Pabit S, Milano S, Aplin C, San Emeterio J, Cerione R, Pollack L
|
| RgGuinier |
2.2 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
27 |
nm3 |
|
|
|
|
|
|
|
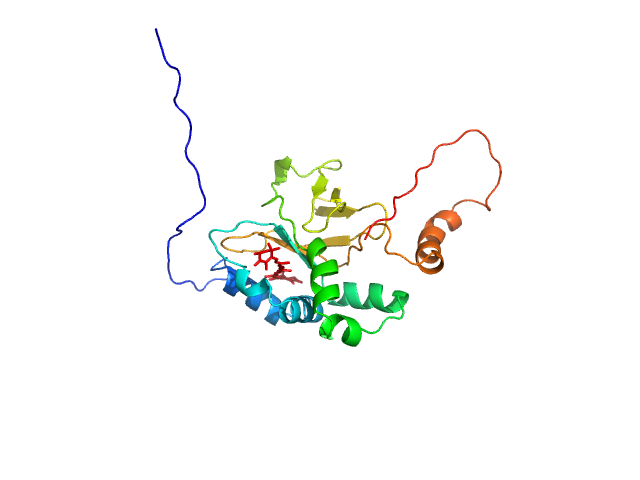
| Sample: |
Astaxanthin binding fasciclin family protein monomer, 22 kDa Coelastrella astaxanthina protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, pH: 7.6 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Sep 24
|
Structural basis for the ligand promiscuity of the neofunctionalized, carotenoid-binding fasciclin domain protein AstaP.
Commun Biol 6(1):471 (2023)
Kornilov FD, Slonimskiy YB, Lunegova DA, Egorkin NA, Savitskaya AG, Kleymenov SY, Maksimov EG, Goncharuk SA, Mineev KS, Sluchanko NN
|
| RgGuinier |
2.1 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
43 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Astaxanthin binding fasciclin family protein monomer, 22 kDa Coelastrella astaxanthina protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, pH: 7.6 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Sep 24
|
Structural basis for the ligand promiscuity of the neofunctionalized, carotenoid-binding fasciclin domain protein AstaP.
Commun Biol 6(1):471 (2023)
Kornilov FD, Slonimskiy YB, Lunegova DA, Egorkin NA, Savitskaya AG, Kleymenov SY, Maksimov EG, Goncharuk SA, Mineev KS, Sluchanko NN
|
| RgGuinier |
2.1 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
46 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
65 kDa invariant surface glycoprotein, putative monomer, 41 kDa Trypanosoma brucei gambiense … protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 3% (v/v) glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2020 Nov 25
|
Cryo-EM structures of Trypanosoma brucei gambiense ISG65 with human complement C3 and C3b and their roles in alternative pathway restriction.
Nat Commun 14(1):2403 (2023)
Sülzen H, Began J, Dhillon A, Kereïche S, Pompach P, Votrubova J, Zahedifard F, Šubrtova A, Šafner M, Hubalek M, Thompson M, Zoltner M, Zoll S
|
| RgGuinier |
3.3 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
53 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
65 kDa invariant surface glycoprotein, putative monomer, 41 kDa Trypanosoma brucei gambiense … protein
Complement C3 monomer, 35 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 3% (v/v) glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2020 Nov 25
|
Cryo-EM structures of Trypanosoma brucei gambiense ISG65 with human complement C3 and C3b and their roles in alternative pathway restriction.
Nat Commun 14(1):2403 (2023)
Sülzen H, Began J, Dhillon A, Kereïche S, Pompach P, Votrubova J, Zahedifard F, Šubrtova A, Šafner M, Hubalek M, Thompson M, Zoltner M, Zoll S
|
| RgGuinier |
3.5 |
nm |
| Dmax |
12.8 |
nm |
| VolumePorod |
104 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Albumin monomer, 66 kDa Bos taurus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at 13A, Taiwan Photon Source, NSRRC on 2022 Jun 7
|
NSRRC TPS13A standard protein archive
Orion Shih
|
| RgGuinier |
2.9 |
nm |
| Dmax |
8.9 |
nm |
| VolumePorod |
84 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Lysozyme C monomer, 14 kDa Gallus gallus protein
|
| Buffer: |
40 mM sodium acetate, 50 mM NaCl, pH: 4 |
| Experiment: |
SAXS
data collected at 13A, Taiwan Photon Source, NSRRC on 2021 Apr 2
|
NSRRC TPS13A standard protein archive
Orion Shih
|
| RgGuinier |
1.4 |
nm |
| Dmax |
4.6 |
nm |
| VolumePorod |
13 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Hemoglobin subunit alpha dimer, 31 kDa Homo sapiens protein
Hemoglobin subunit beta dimer, 32 kDa Homo sapiens protein
|
| Buffer: |
phosphate buffered saline, pH: 7.5 |
| Experiment: |
SAXS
data collected at 13A, Taiwan Photon Source, NSRRC on 2021 Oct 7
|
NSRRC TPS13A standard protein archive
Orion Shih
|
| RgGuinier |
2.5 |
nm |
| Dmax |
9.2 |
nm |
| VolumePorod |
97 |
nm3 |
|
|
|
|
|
|
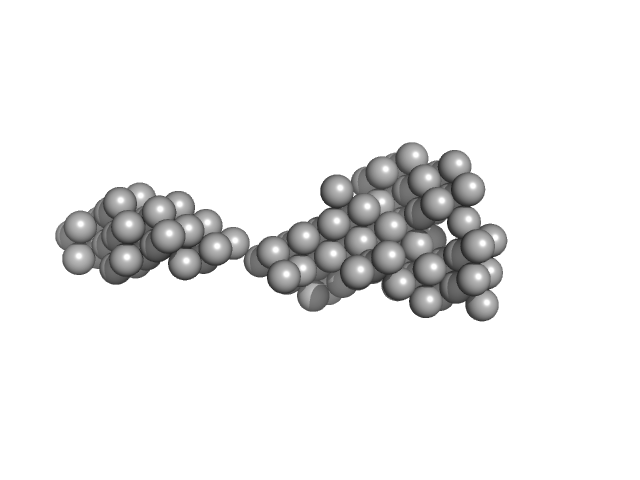
|
| Sample: |
Human Mucin 2 C-terminal dimer, 240 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 100 mM NaCl, 10 mM CaCl2, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2020 Nov 20
|
The intestinal MUC2 mucin C-terminus is stabilized by an extra disulfide bond in comparison to von Willebrand factor and other gel-forming mucins
Nature Communications 14(1) (2023)
Gallego P, Garcia-Bonete M, Trillo-Muyo S, Recktenwald C, Johansson M, Hansson G
|
| RgGuinier |
8.4 |
nm |
| Dmax |
32.0 |
nm |
| VolumePorod |
684 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Angiotensin-converting enzyme 2 chimera with the Fc region of the immunoglobulin heavy constant gamma 4 dimer, 217 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, SFB 1035, Technische Universität München on 2021 Jul 5
|
Extrinsic stabilization of antiviral ACE2-Fc fusion proteins targeting SARS-CoV-2.
Commun Biol 6(1):386 (2023)
Svilenov HL, Delhommel F, Siebenmorgen T, Rührnößl F, Popowicz GM, Reiter A, Sattler M, Brockmeyer C, Buchner J
|
| RgGuinier |
5.5 |
nm |
| Dmax |
18.6 |
nm |
| VolumePorod |
408 |
nm3 |
|
|