|
|
|
|
|
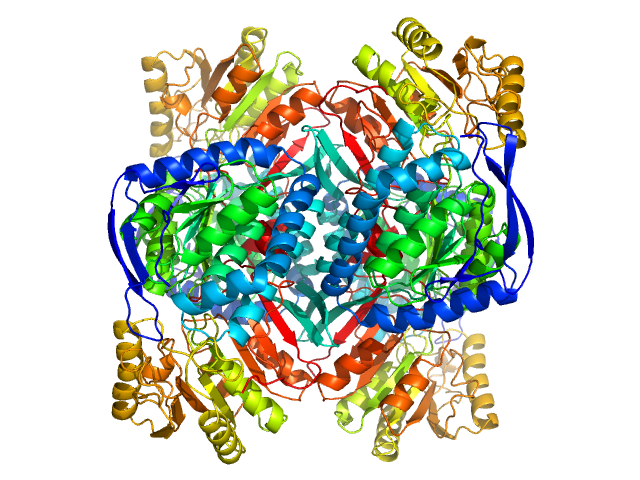
| Sample: |
4-trimethylaminobutyraldehyde dehydrogenase tetramer, 215 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 600 mM NaCl, 2% (v/v) glycerol, 1 mM DTT, 1 mM NAD+, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 May 28
|
Inhibition, crystal structures, and in-solution oligomeric structure of aldehyde dehydrogenase 9A1.
Arch Biochem Biophys :108477 (2020)
Wyatt JW, Korasick DA, Qureshi IA, Campbell AC, Gates KS, Tanner JJ
|
| RgGuinier |
3.8 |
nm |
| Dmax |
10.9 |
nm |
| VolumePorod |
252 |
nm3 |
|
|
|
|
|
|
|
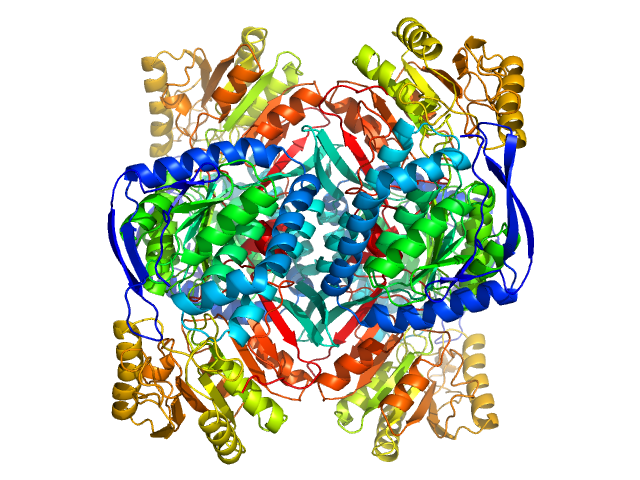
| Sample: |
4-trimethylaminobutyraldehyde dehydrogenase tetramer, 215 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 600 mM NaCl, 2% (v/v) glycerol, 1 mM DTT, 1 mM NAD+, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 May 28
|
Inhibition, crystal structures, and in-solution oligomeric structure of aldehyde dehydrogenase 9A1.
Arch Biochem Biophys :108477 (2020)
Wyatt JW, Korasick DA, Qureshi IA, Campbell AC, Gates KS, Tanner JJ
|
| RgGuinier |
3.8 |
nm |
| Dmax |
10.8 |
nm |
| VolumePorod |
240 |
nm3 |
|
|
|
|
|
|
|
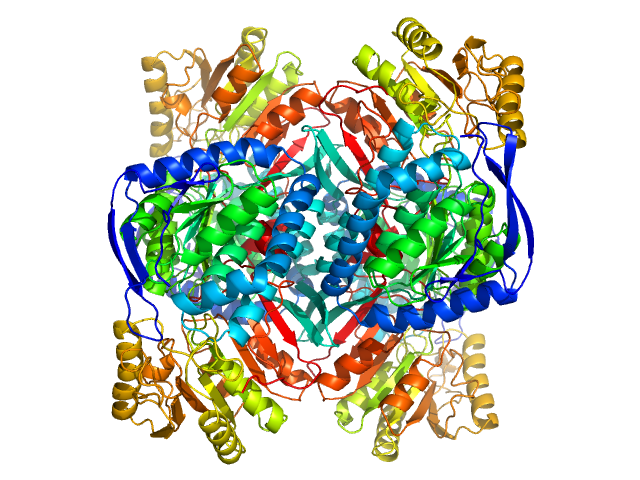
| Sample: |
4-trimethylaminobutyraldehyde dehydrogenase tetramer, 215 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 600 mM NaCl, 2% (v/v) glycerol, 1 mM DTT, 1 mM NAD+, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 May 28
|
Inhibition, crystal structures, and in-solution oligomeric structure of aldehyde dehydrogenase 9A1.
Arch Biochem Biophys :108477 (2020)
Wyatt JW, Korasick DA, Qureshi IA, Campbell AC, Gates KS, Tanner JJ
|
| RgGuinier |
3.8 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
240 |
nm3 |
|
|
|
|
|
|
|
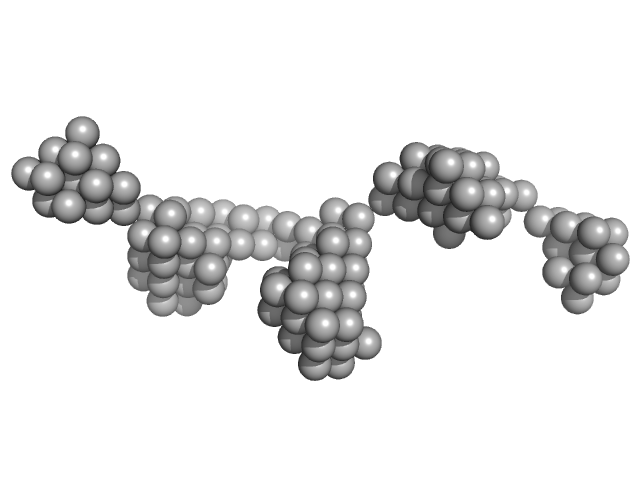
| Sample: |
Beta-ketoacyl synthase Bamb_5925 monomer, 4 kDa Burkholderia ambifaria protein
|
| Buffer: |
20 mM Tris-HCl, 200 mM NaCl, 5% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Jan 23
|
Towards improved understanding of intersubunit interactions in modular polyketide biosynthesis: docking in the enacyloxin IIa polyketide synthase.
J Struct Biol :107581 (2020)
Risser F, Collin S, Dos Santos-Morais R, Gruez A, Chagot B, Weissman KJ
|
| RgGuinier |
1.6 |
nm |
| Dmax |
6.2 |
nm |
| VolumePorod |
3 |
nm3 |
|
|
|
|
|
|

|
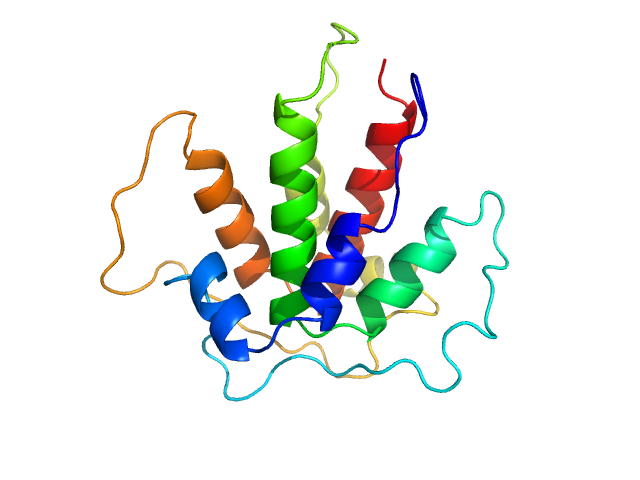
| Sample: |
Beta-ketoacyl synthase Bamb_5924 dimer, 10 kDa Burkholderia ambifaria protein
|
| Buffer: |
20 mM Tris-HCl, 200 mM NaCl, 5% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2017 Oct 6
|
Towards improved understanding of intersubunit interactions in modular polyketide biosynthesis: docking in the enacyloxin IIa polyketide synthase.
J Struct Biol :107581 (2020)
Risser F, Collin S, Dos Santos-Morais R, Gruez A, Chagot B, Weissman KJ
|
| RgGuinier |
1.9 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
22 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Beta-ketoacyl synthase Bamb_5920 monomer, 2 kDa Burkholderia ambifaria protein
|
| Buffer: |
20 mM Tris-HCl, 200 mM NaCl, 5% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Jan 23
|
Towards improved understanding of intersubunit interactions in modular polyketide biosynthesis: docking in the enacyloxin IIa polyketide synthase.
J Struct Biol :107581 (2020)
Risser F, Collin S, Dos Santos-Morais R, Gruez A, Chagot B, Weissman KJ
|
| RgGuinier |
1.2 |
nm |
| Dmax |
4.9 |
nm |
| VolumePorod |
3 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Beta-ketoacyl synthase Bamb_5924, K32A mutant dimer, 10 kDa Burkholderia ambifaria protein
|
| Buffer: |
20 mM Tris-HCl, 200 mM NaCl, 5% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Jan 23
|
Towards improved understanding of intersubunit interactions in modular polyketide biosynthesis: docking in the enacyloxin IIa polyketide synthase.
J Struct Biol :107581 (2020)
Risser F, Collin S, Dos Santos-Morais R, Gruez A, Chagot B, Weissman KJ
|
| RgGuinier |
1.8 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
22 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Beta-ketoacyl synthase Bamb_5924, R17A mutant dimer, 10 kDa Burkholderia ambifaria protein
|
| Buffer: |
20 mM Tris-HCl, 200 mM NaCl, 5% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Jan 23
|
Towards improved understanding of intersubunit interactions in modular polyketide biosynthesis: docking in the enacyloxin IIa polyketide synthase.
J Struct Biol :107581 (2020)
Risser F, Collin S, Dos Santos-Morais R, Gruez A, Chagot B, Weissman KJ
|
| RgGuinier |
1.8 |
nm |
| Dmax |
8.2 |
nm |
| VolumePorod |
20 |
nm3 |
|
|
|
|
|
|

|
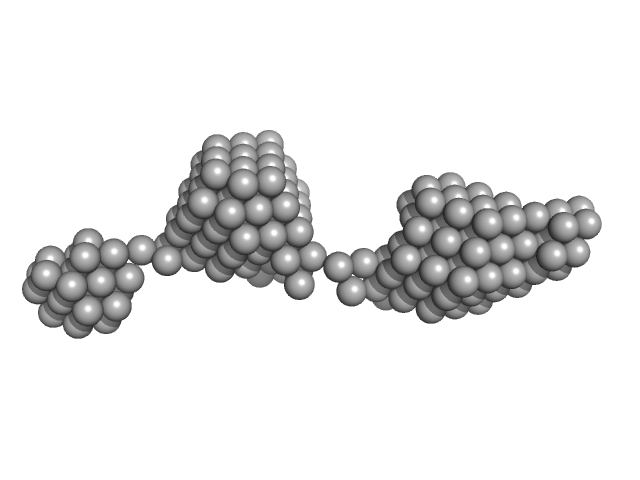
| Sample: |
Beta-ketoacyl synthase Bamb_5920 CDD/Bamb_5919 NDD artificial protein fusion dimer, 13 kDa Burkholderia ambifaria protein
|
| Buffer: |
20 mM Tris-HCl, 200 mM NaCl, 5% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Jan 23
|
Towards improved understanding of intersubunit interactions in modular polyketide biosynthesis: docking in the enacyloxin IIa polyketide synthase.
J Struct Biol :107581 (2020)
Risser F, Collin S, Dos Santos-Morais R, Gruez A, Chagot B, Weissman KJ
|
| RgGuinier |
1.8 |
nm |
| Dmax |
6.0 |
nm |
| VolumePorod |
25 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Beta-ketoacyl synthase Bamb_5925 CDD/Bamb_5924 NDD artificial protein fusion dimer, 18 kDa Burkholderia ambifaria protein
|
| Buffer: |
20 mM Tris-HCl, 200 mM NaCl, 5% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Jan 23
|
Towards improved understanding of intersubunit interactions in modular polyketide biosynthesis: docking in the enacyloxin IIa polyketide synthase.
J Struct Biol :107581 (2020)
Risser F, Collin S, Dos Santos-Morais R, Gruez A, Chagot B, Weissman KJ
|
| RgGuinier |
2.1 |
nm |
| Dmax |
9.6 |
nm |
| VolumePorod |
47 |
nm3 |
|
|