|
|
|
|
|
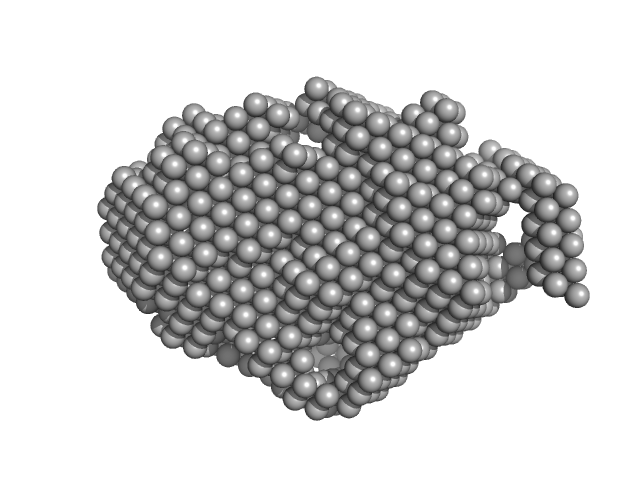
| Sample: |
Trehalose transferase (Trehalose phosphorylase/synthase) monomer, 47 kDa Thermoproteus uzoniensis protein
|
| Buffer: |
50 mM HEPES, 100 NaCl, 4 mM DTT,, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Sep 27
|
Anomeric Selectivity of Trehalose Transferase with Rare L-Sugars
ACS Catalysis (2020)
Mestrom L, Marsden S, van der Eijk H, Laustsen J, Jeffries C, Svergun D, Hagedoorn P, Bento I, Hanefeld U
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.8 |
nm |
| VolumePorod |
77 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Trehalose transferase (Trehalose phosphorylase/synthase) monomer, 47 kDa Thermoproteus uzoniensis protein
|
| Buffer: |
50 mM HEPES, 100 NaCl, 4 mM DTT, 20 mM MgCl2, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Sep 27
|
Anomeric Selectivity of Trehalose Transferase with Rare L-Sugars
ACS Catalysis (2020)
Mestrom L, Marsden S, van der Eijk H, Laustsen J, Jeffries C, Svergun D, Hagedoorn P, Bento I, Hanefeld U
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
76 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Trehalose transferase (Trehalose phosphorylase/synthase) monomer, 47 kDa Thermoproteus uzoniensis protein
|
| Buffer: |
50 mM HEPES, 100 NaCl, 4 mM DTT, 20 mM MgCl2, 1 mM trehalose, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Sep 27
|
Anomeric Selectivity of Trehalose Transferase with Rare L-Sugars
ACS Catalysis (2020)
Mestrom L, Marsden S, van der Eijk H, Laustsen J, Jeffries C, Svergun D, Hagedoorn P, Bento I, Hanefeld U
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.3 |
nm |
| VolumePorod |
76 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Trehalose transferase (Trehalose phosphorylase/synthase) monomer, 47 kDa Thermoproteus uzoniensis protein
|
| Buffer: |
50 mM HEPES, 100 NaCl, 4 mM DTT, 20 mM MgCl2, 1 mM UDP-glucose, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Sep 27
|
Anomeric Selectivity of Trehalose Transferase with Rare L-Sugars
ACS Catalysis (2020)
Mestrom L, Marsden S, van der Eijk H, Laustsen J, Jeffries C, Svergun D, Hagedoorn P, Bento I, Hanefeld U
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
73 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Collagen, type VI, alpha 3 monomer, 44 kDa Mus musculus protein
|
| Buffer: |
20 mM TRIS, 150mM NaCl 3% v/v glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Apr 20
|
Structure of a collagen VI α3 chain VWA domain array: adaptability and functional implications of myopathy causing mutations
Journal of Biological Chemistry :jbc.RA120.014865 (2020)
Solomon-Degefa H, Gebauer J, Jeffries C, Freiburg C, Meckelburg P, Bird L, Baumann U, Svergun D, Owens R, Werner J, Behrmann E, Paulsson M, Wagener R
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.9 |
nm |
| VolumePorod |
66 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Collagen, type VI, alpha 3 monomer, 93 kDa Mus musculus protein
|
| Buffer: |
20 mM TRIS, 150mM NaCl 3% v/v glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Nov 4
|
Structure of a collagen VI α3 chain VWA domain array: adaptability and functional implications of myopathy causing mutations
Journal of Biological Chemistry :jbc.RA120.014865 (2020)
Solomon-Degefa H, Gebauer J, Jeffries C, Freiburg C, Meckelburg P, Bird L, Baumann U, Svergun D, Owens R, Werner J, Behrmann E, Paulsson M, Wagener R
|
| RgGuinier |
4.1 |
nm |
| Dmax |
15.6 |
nm |
| VolumePorod |
143 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Upstream of N-ras, isoform A monomer, 29 kDa Drosophila melanogaster protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Sep 15
|
Pseudo-RNA-Binding Domains Mediate RNA Structure Specificity in Upstream of N-Ras.
Cell Rep 32(3):107930 (2020)
Hollmann NM, Jagtap PKA, Masiewicz P, Guitart T, Simon B, Provaznik J, Stein F, Haberkant P, Sweetapple LJ, Villacorta L, Mooijman D, Benes V, Savitski MM, Gebauer F, Hennig J
|
| RgGuinier |
2.7 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
38 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Upstream of N-ras, isoform A monomer, 19 kDa Drosophila melanogaster protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 May 14
|
Pseudo-RNA-Binding Domains Mediate RNA Structure Specificity in Upstream of N-Ras.
Cell Rep 32(3):107930 (2020)
Hollmann NM, Jagtap PKA, Masiewicz P, Guitart T, Simon B, Provaznik J, Stein F, Haberkant P, Sweetapple LJ, Villacorta L, Mooijman D, Benes V, Savitski MM, Gebauer F, Hennig J
|
| RgGuinier |
1.8 |
nm |
| Dmax |
6.0 |
nm |
| VolumePorod |
28 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Collagen alpha-3(VI) chain, N2 domain monomer, 22 kDa Homo sapiens protein
|
| Buffer: |
20 mM TRIS, pH 7.4, 150mM NaCl 3% v/v glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 May 4
|
Structure of a collagen VI α3 chain VWA domain array: adaptability and functional implications of myopathy causing mutations
Journal of Biological Chemistry :jbc.RA120.014865 (2020)
Solomon-Degefa H, Gebauer J, Jeffries C, Freiburg C, Meckelburg P, Bird L, Baumann U, Svergun D, Owens R, Werner J, Behrmann E, Paulsson M, Wagener R
|
| RgGuinier |
1.8 |
nm |
| Dmax |
5.8 |
nm |
| VolumePorod |
40 |
nm3 |
|
|
|
|
|
|
|
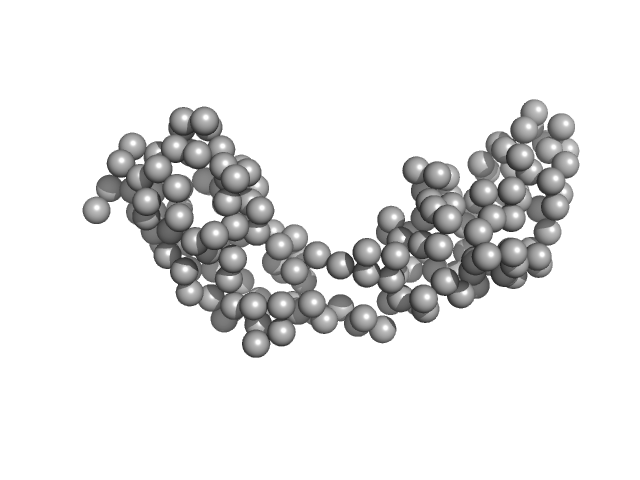
| Sample: |
Minimal hepatocyte growth factor mimic K1K1 monomer, 19 kDa synthetic construct protein
|
| Buffer: |
25 mM Tris, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Nov 3
|
Dimerization of kringle 1 domain from hepatocyte growth factor/scatter factor provides a potent MET receptor agonist
Life Science Alliance 5(12):e202201424 (2022)
de Nola G, Leclercq B, Mougel A, Taront S, Simonneau C, Forneris F, Adriaenssens E, Drobecq H, Iamele L, Dubuquoy L, Melnyk O, Gherardi E, de Jonge H, Vicogne J
|
| RgGuinier |
2.2 |
nm |
| Dmax |
6.6 |
nm |
| VolumePorod |
22 |
nm3 |
|
|