|
|
|
|
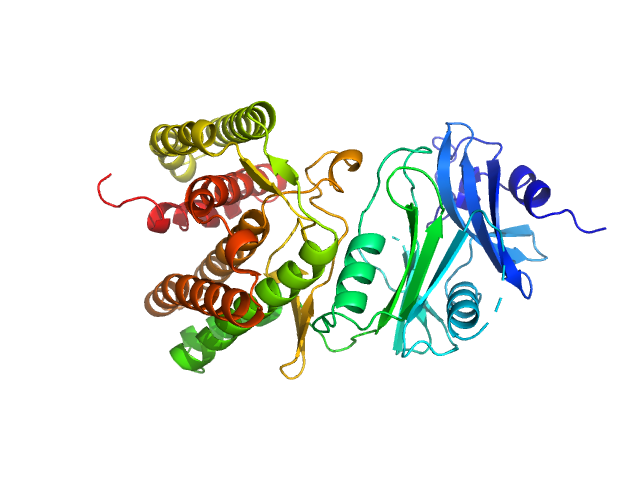
|
| Sample: |
Glucosamine kinase monomer, 48 kDa Streptacidiphilus jiangxiensis protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 10 mM MgCl2, 5 mM DTT, 1 mM ATP, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Nov 24
|
Molecular Fingerprints for a Novel Enzyme Family in Actinobacteria with Glucosamine Kinase Activity.
MBio 10(3) (2019)
Manso JA, Nunes-Costa D, Macedo-Ribeiro S, Empadinhas N, Pereira PJB
|
| RgGuinier |
2.5 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
73 |
nm3 |
|
|
|
|
|
|
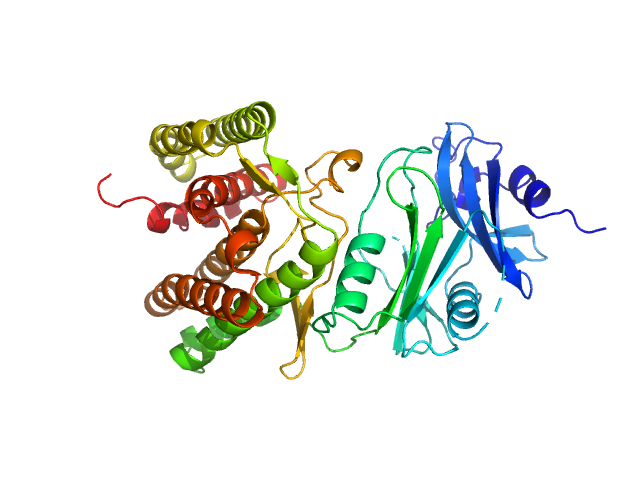
|
| Sample: |
Glucosamine kinase monomer, 48 kDa Streptacidiphilus jiangxiensis protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 10 mM MgCl2, 5 mM DTT, 0.2 M D-glucosamine, 1 mM ATP, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Nov 20
|
Molecular Fingerprints for a Novel Enzyme Family in Actinobacteria with Glucosamine Kinase Activity.
MBio 10(3) (2019)
Manso JA, Nunes-Costa D, Macedo-Ribeiro S, Empadinhas N, Pereira PJB
|
| RgGuinier |
2.5 |
nm |
| Dmax |
7.7 |
nm |
| VolumePorod |
76 |
nm3 |
|
|
|
|
|
|
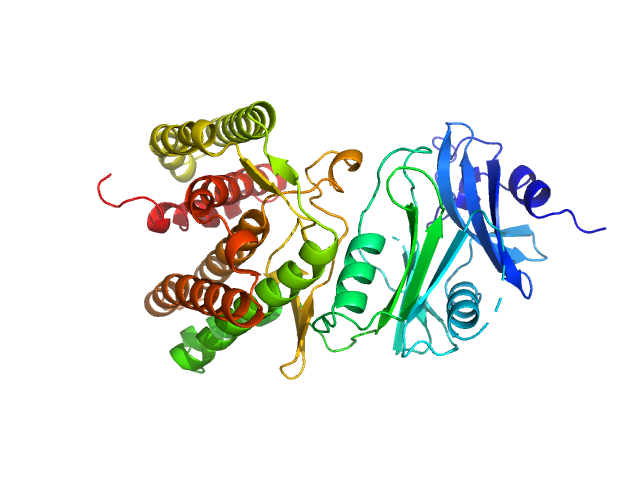
|
| Sample: |
Glucosamine kinase monomer, 48 kDa Streptacidiphilus jiangxiensis protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 10 mM MgCl2, 5 mM DTT, 50 mM D-glucose, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Apr 13
|
Molecular Fingerprints for a Novel Enzyme Family in Actinobacteria with Glucosamine Kinase Activity.
MBio 10(3) (2019)
Manso JA, Nunes-Costa D, Macedo-Ribeiro S, Empadinhas N, Pereira PJB
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
73 |
nm3 |
|
|
|
|
|
|
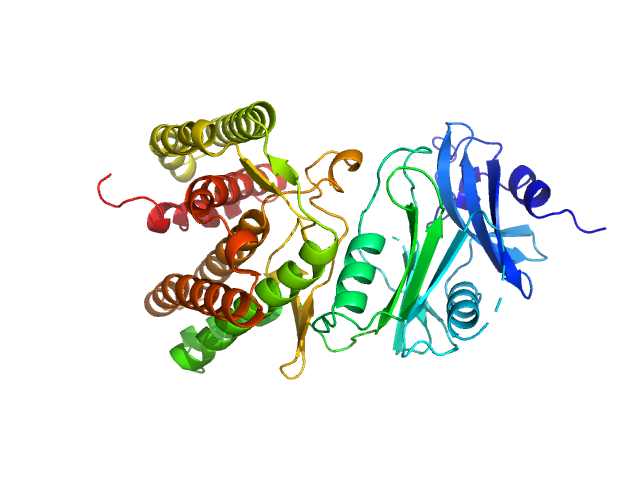
|
| Sample: |
Glucosamine kinase monomer, 48 kDa Streptacidiphilus jiangxiensis protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 10 mM MgCl2, 5 mM DTT, 50 mM D-glucose, 1 mM ATP, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Nov 24
|
Molecular Fingerprints for a Novel Enzyme Family in Actinobacteria with Glucosamine Kinase Activity.
MBio 10(3) (2019)
Manso JA, Nunes-Costa D, Macedo-Ribeiro S, Empadinhas N, Pereira PJB
|
| RgGuinier |
2.5 |
nm |
| Dmax |
7.7 |
nm |
| VolumePorod |
73 |
nm3 |
|
|
|
|
|
|
|
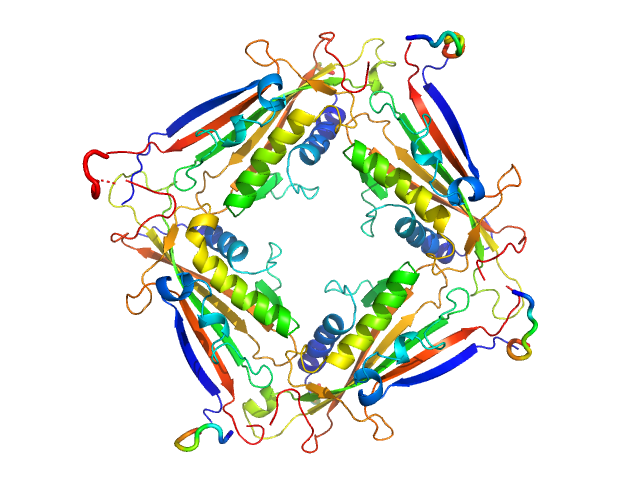
| Sample: |
Transient receptor potential channel mucolipin 2 tetramer, 93 kDa Homo sapiens protein
|
| Buffer: |
10 mM HEPES pH 7.4, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Oct 18
|
Structure of the Human TRPML2 Ion Channel Extracytosolic/Lumenal Domain.
Structure (2019)
Viet KK, Wagner A, Schwickert K, Hellwig N, Brennich M, Bader N, Schirmeister T, Morgner N, Schindelin H, Hellmich UA
|
| RgGuinier |
3.4 |
nm |
| Dmax |
8.9 |
nm |
| VolumePorod |
134 |
nm3 |
|
|
|
|
|
|
|
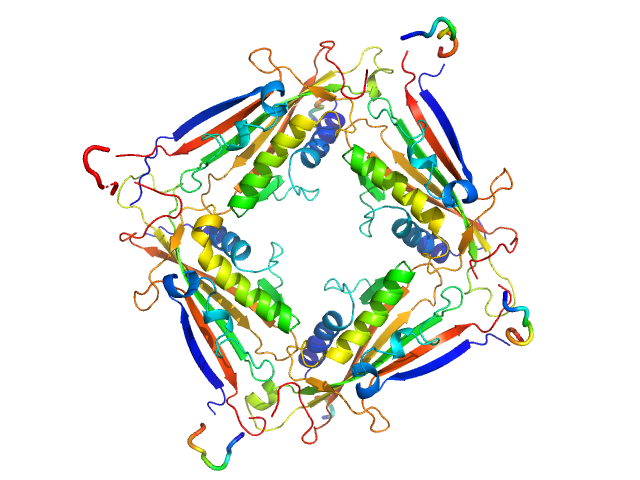
| Sample: |
Transient receptor potential channel mucolipin 2 tetramer, 93 kDa Homo sapiens protein
|
| Buffer: |
10 mM HEPES pH 7.4, 150 mM NaCl, 2 mM CaCl2, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Oct 20
|
Structure of the Human TRPML2 Ion Channel Extracytosolic/Lumenal Domain.
Structure (2019)
Viet KK, Wagner A, Schwickert K, Hellwig N, Brennich M, Bader N, Schirmeister T, Morgner N, Schindelin H, Hellmich UA
|
| RgGuinier |
3.4 |
nm |
| Dmax |
8.9 |
nm |
| VolumePorod |
134 |
nm3 |
|
|
|
|
|
|
|
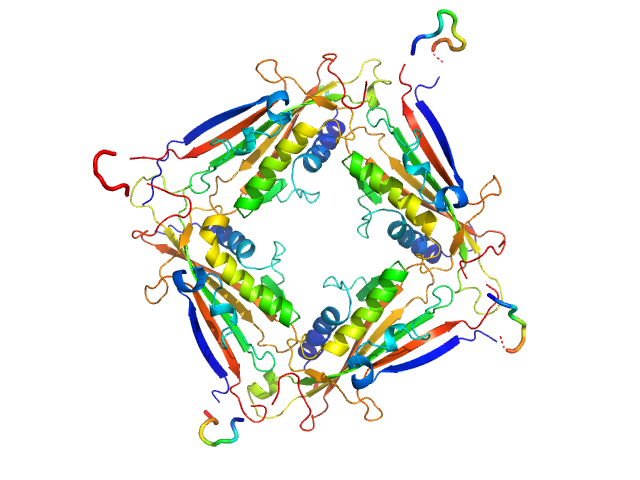
| Sample: |
Transient receptor potential channel mucolipin 2 tetramer, 93 kDa Homo sapiens protein
|
| Buffer: |
10 mM Hepes, pH 6.5, 150 mM NaCl, 0.5 mM CaCl2, pH: 6.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Oct 20
|
Structure of the Human TRPML2 Ion Channel Extracytosolic/Lumenal Domain.
Structure (2019)
Viet KK, Wagner A, Schwickert K, Hellwig N, Brennich M, Bader N, Schirmeister T, Morgner N, Schindelin H, Hellmich UA
|
| RgGuinier |
3.4 |
nm |
| Dmax |
8.9 |
nm |
| VolumePorod |
132 |
nm3 |
|
|
|
|
|
|
|
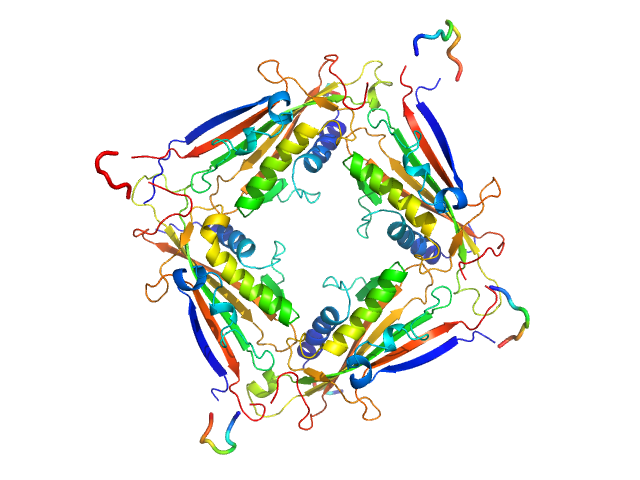
| Sample: |
Transient receptor potential channel mucolipin 2 tetramer, 93 kDa Homo sapiens protein
|
| Buffer: |
10 mM HEPES pH 6.5 150 mM NaCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Oct 18
|
Structure of the Human TRPML2 Ion Channel Extracytosolic/Lumenal Domain.
Structure (2019)
Viet KK, Wagner A, Schwickert K, Hellwig N, Brennich M, Bader N, Schirmeister T, Morgner N, Schindelin H, Hellmich UA
|
| RgGuinier |
3.4 |
nm |
| Dmax |
8.9 |
nm |
| VolumePorod |
140 |
nm3 |
|
|
|
|
|
|
|
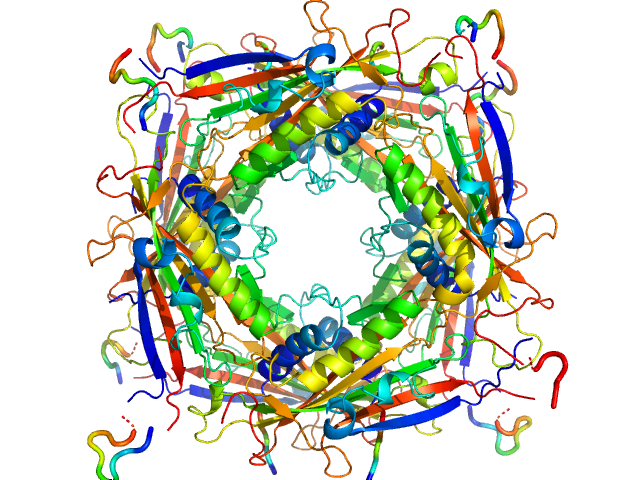
| Sample: |
Transient receptor potential channel mucolipin 2 octamer, 187 kDa Homo sapiens protein
|
| Buffer: |
10 mM Hepes, pH 4.5, 150 mM NaCl, pH: 4.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Oct 18
|
Structure of the Human TRPML2 Ion Channel Extracytosolic/Lumenal Domain.
Structure (2019)
Viet KK, Wagner A, Schwickert K, Hellwig N, Brennich M, Bader N, Schirmeister T, Morgner N, Schindelin H, Hellmich UA
|
| RgGuinier |
3.7 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
285 |
nm3 |
|
|
|
|
|
|
|
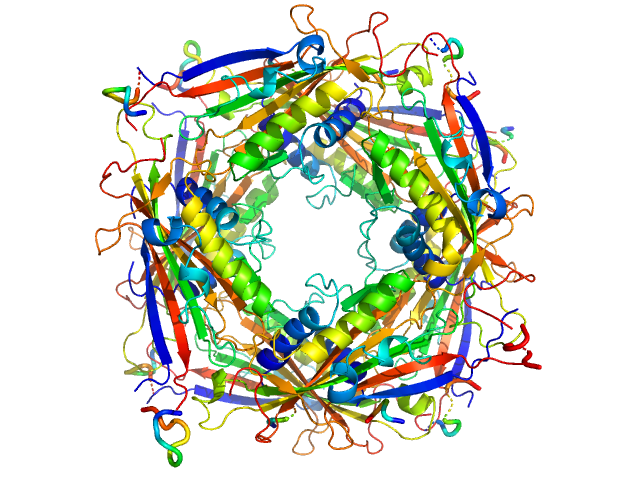
| Sample: |
Transient receptor potential channel mucolipin 2 octamer, 187 kDa Homo sapiens protein
|
| Buffer: |
10 mM Hepes, pH 4.5, 150 mM NaCl, 0.5 mM CaCl2, pH: 4.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Oct 20
|
Structure of the Human TRPML2 Ion Channel Extracytosolic/Lumenal Domain.
Structure (2019)
Viet KK, Wagner A, Schwickert K, Hellwig N, Brennich M, Bader N, Schirmeister T, Morgner N, Schindelin H, Hellmich UA
|
| RgGuinier |
3.7 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
280 |
nm3 |
|
|