|
|
|
|
|
| Sample: |
Microtubule-associated protein 2, isoform 3 monomer, 49 kDa Rattus norvegicus protein
|
| Buffer: |
50 mM MOPS, 150 mM NaCl, 0.03% NaN3, pH: 6.9 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Apr 21
|
Functionally specific binding regions of microtubule-associated protein 2c exhibit distinct conformations and dynamics.
J Biol Chem 293(34):13297-13309 (2018)
Melková K, Zapletal V, Jansen S, Nomilner E, Zachrdla M, Hritz J, Nováček J, Zweckstetter M, Jensen MR, Blackledge M, Žídek L
|
|
|
|
|
|
|
|
| Sample: |
Microtubule-associated protein 2, isoform 3 monomer, 49 kDa Rattus norvegicus protein
|
| Buffer: |
MOPS map2c buffer, pH: 6.9 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Apr 21
|
Functionally specific binding regions of microtubule-associated protein 2c exhibit distinct conformations and dynamics.
J Biol Chem 293(34):13297-13309 (2018)
Melková K, Zapletal V, Jansen S, Nomilner E, Zachrdla M, Hritz J, Nováček J, Zweckstetter M, Jensen MR, Blackledge M, Žídek L
|
|
|
|
|
|
|

|
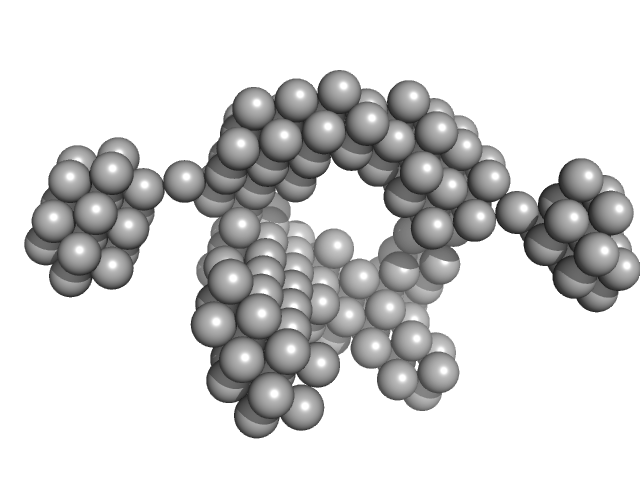
| Sample: |
VNG0258H/RosR dimer, 29 kDa Halobacterium salinarum NRC-1 protein
|
| Buffer: |
50 mM HEPES, 2 M KCl, 0.02% NaN3, pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Mar 7
|
The 3-D structure of VNG0258H/RosR - A haloarchaeal DNA-binding protein in its ionic shell.
J Struct Biol (2018)
Kutnowski N, Shmuely H, Dahan I, Shmulevich F, Davidov G, Shahar A, Eichler J, Zarivach R, Shaanan B
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
58 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
VNG0258H/RosR dimer, 29 kDa Halobacterium salinarum NRC-1 protein
|
| Buffer: |
50 mM HEPES, 2 M NaCl, 0.02% NaN3, pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Mar 7
|
The 3-D structure of VNG0258H/RosR - A haloarchaeal DNA-binding protein in its ionic shell.
J Struct Biol (2018)
Kutnowski N, Shmuely H, Dahan I, Shmulevich F, Davidov G, Shahar A, Eichler J, Zarivach R, Shaanan B
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.7 |
nm |
| VolumePorod |
63 |
nm3 |
|
|
|
|
|
|
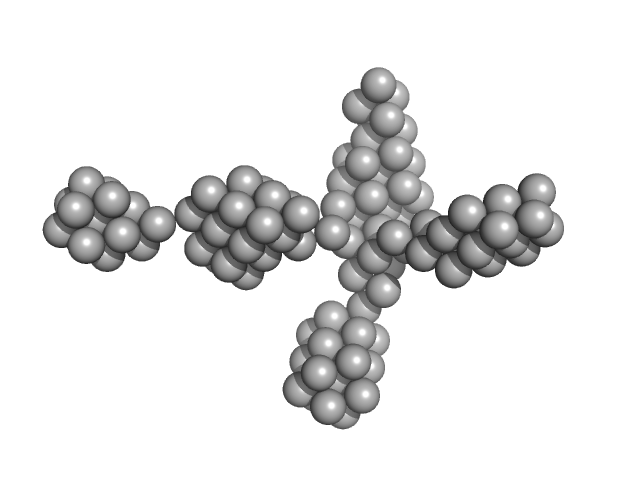
|
| Sample: |
VNG0258H/RosR dimer, 29 kDa Halobacterium salinarum NRC-1 protein
|
| Buffer: |
50 mM HEPES, 2 M KBr, 0.02% NaN3, pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Mar 7
|
The 3-D structure of VNG0258H/RosR - A haloarchaeal DNA-binding protein in its ionic shell.
J Struct Biol (2018)
Kutnowski N, Shmuely H, Dahan I, Shmulevich F, Davidov G, Shahar A, Eichler J, Zarivach R, Shaanan B
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.7 |
nm |
| VolumePorod |
46 |
nm3 |
|
|
|
|
|
|
|
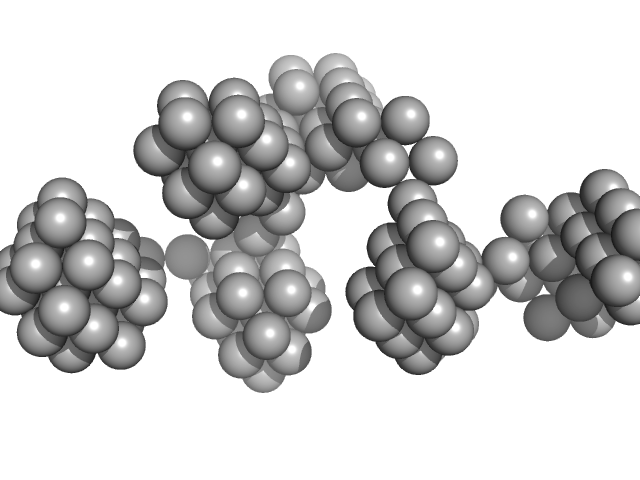
| Sample: |
VNG0258H/RosR dimer, 29 kDa Halobacterium salinarum NRC-1 protein
|
| Buffer: |
50 mM HEPES, 2 M NaBr, 0.02% NaN3, pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Sep 26
|
The 3-D structure of VNG0258H/RosR - A haloarchaeal DNA-binding protein in its ionic shell.
J Struct Biol (2018)
Kutnowski N, Shmuely H, Dahan I, Shmulevich F, Davidov G, Shahar A, Eichler J, Zarivach R, Shaanan B
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.1 |
nm |
| VolumePorod |
59 |
nm3 |
|
|
|
|
|
|
|
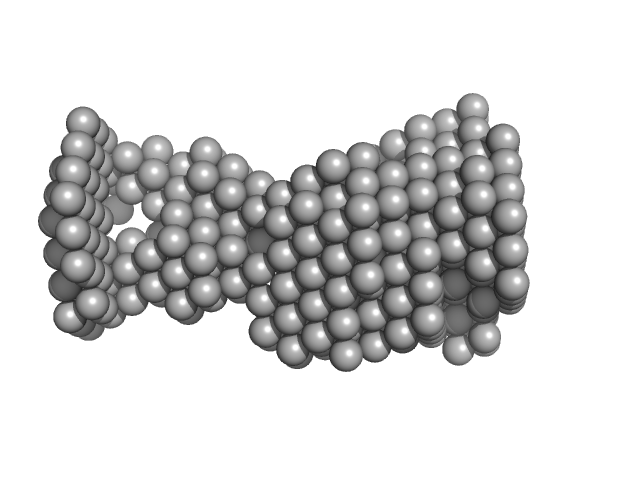
| Sample: |
VNG0258H/RosR dimer, 29 kDa Halobacterium salinarum NRC-1 protein
|
| Buffer: |
50 mM HEPES, 2 M RbCl, 0.02% NaN3, pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Mar 7
|
The 3-D structure of VNG0258H/RosR - A haloarchaeal DNA-binding protein in its ionic shell.
J Struct Biol (2018)
Kutnowski N, Shmuely H, Dahan I, Shmulevich F, Davidov G, Shahar A, Eichler J, Zarivach R, Shaanan B
|
| RgGuinier |
3.3 |
nm |
| Dmax |
9.3 |
nm |
| VolumePorod |
89 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Procollagen lysyl hydroxylase LH3 dimer, 180 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 200 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Feb 28
|
Molecular architecture of the multifunctional collagen lysyl hydroxylase and glycosyltransferase LH3.
Nat Commun 9(1):3163 (2018)
Scietti L, Chiapparino A, De Giorgi F, Fumagalli M, Khoriauli L, Nergadze S, Basu S, Olieric V, Cucca L, Banushi B, Profumo A, Giulotto E, Gissen P, Forneris F
|
| RgGuinier |
5.1 |
nm |
| Dmax |
21.0 |
nm |
| VolumePorod |
268 |
nm3 |
|
|
|
|
|
|
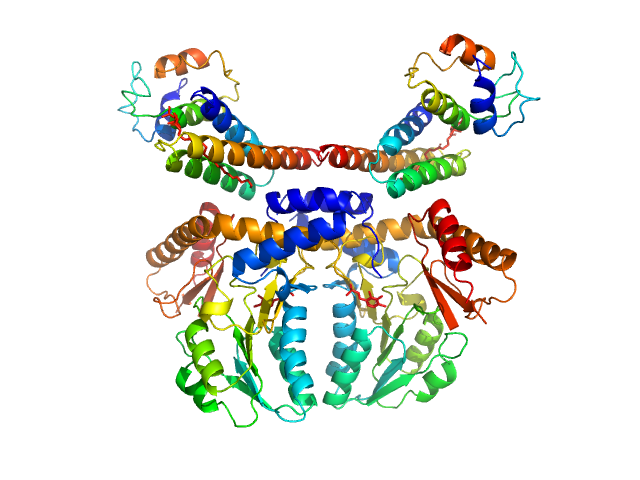
|
| Sample: |
Cysteine desulfurase, mitochondrial dimer, 90 kDa Homo sapiens protein
LYR motif-containing protein 4 dimer, 23 kDa Homo sapiens protein
Acyl carrier protein dimer, 22 kDa Escherichia coli protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at Bruker Nanostar, NMRFAM on 2017 May 22
|
Architectural Features of Human Mitochondrial Cysteine Desulfurase Complexes from Crosslinking Mass Spectrometry and Small-Angle X-Ray Scattering.
Structure 26(8):1127-1136.e4 (2018)
Cai K, Frederick RO, Dashti H, Markley JL
|
| RgGuinier |
3.7 |
nm |
| Dmax |
11.8 |
nm |
| VolumePorod |
204 |
nm3 |
|
|
|
|
|
|
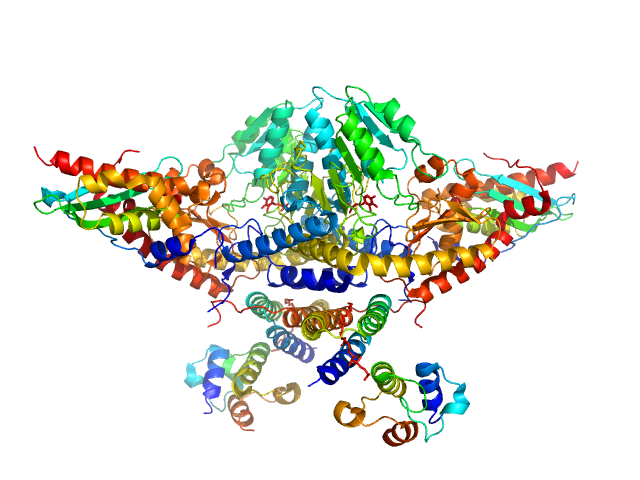
|
| Sample: |
Cysteine desulfurase, mitochondrial dimer, 90 kDa Homo sapiens protein
LYR motif-containing protein 4 dimer, 23 kDa Homo sapiens protein
Acyl carrier protein dimer, 22 kDa Escherichia coli protein
Iron-sulfur cluster assembly enzyme ISCU, mitochondrial dimer, 29 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at Bruker Nanostar, NMRFAM on 2017 May 22
|
Architectural Features of Human Mitochondrial Cysteine Desulfurase Complexes from Crosslinking Mass Spectrometry and Small-Angle X-Ray Scattering.
Structure 26(8):1127-1136.e4 (2018)
Cai K, Frederick RO, Dashti H, Markley JL
|
| RgGuinier |
3.9 |
nm |
| Dmax |
13.7 |
nm |
| VolumePorod |
218 |
nm3 |
|
|