|
|
|
|

|
| Sample: |
S-crystallin, Q25367_DOROP dimer, 55 kDa Doryteuthis pealeii protein
|
| Buffer: |
Tissue fluid, pH: 7 |
| Experiment: |
SAXS
data collected at X9A, National Synchrotron Light Source (NSLS) on 2014 Jul 17
|
Eye patches: Protein assembly of index-gradient squid lenses.
Science 357(6351):564-569 (2017)
Cai J, Townsend JP, Dodson TC, Heiney PA, Sweeney AM
|
|
|
|
|
|
|

|
| Sample: |
S-crystallin, Q25367_DOROP dimer, 55 kDa Doryteuthis pealeii protein
|
| Buffer: |
Tissue fluid, pH: 7 |
| Experiment: |
SAXS
data collected at X9A, National Synchrotron Light Source (NSLS) on 2014 Jul 17
|
Eye patches: Protein assembly of index-gradient squid lenses.
Science 357(6351):564-569 (2017)
Cai J, Townsend JP, Dodson TC, Heiney PA, Sweeney AM
|
|
|
|
|
|
|
|
| Sample: |
S-crystallin, Q25367_DOROP dimer, 55 kDa Doryteuthis pealeii protein
|
| Buffer: |
PBS, phosphate buffered saline, pH: 7 |
| Experiment: |
SAXS
data collected at X9A, National Synchrotron Light Source (NSLS) on 2013 Nov 12
|
Eye patches: Protein assembly of index-gradient squid lenses.
Science 357(6351):564-569 (2017)
Cai J, Townsend JP, Dodson TC, Heiney PA, Sweeney AM
|
|
|
|
|
|
|
|
| Sample: |
Bovine β Cardiac Myosin S1 fragment monomer, 117 kDa Bos taurus protein
|
| Buffer: |
10 mM HEPES, 50 mM NaCl, 1 mM NaN3, 2.5 mM MgCl2, 2 mM ADP, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2016 Mar 1
|
Mechanistic and structural basis for activation of cardiac myosin force production by omecamtiv mecarbil.
Nat Commun 8(1):190 (2017)
Planelles-Herrero VJ, Hartman JJ, Robert-Paganin J, Malik FI, Houdusse A
|
| RgGuinier |
3.6 |
nm |
| Dmax |
12.6 |
nm |
| VolumePorod |
162 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Bovine β Cardiac Myosin S1 fragment monomer, 117 kDa Bos taurus protein
|
| Buffer: |
10 mM HEPES, 50 mM NaCl, 1 mM NaN3, 2.5 mM MgCl2, 2 mM ADP, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2016 Mar 1
|
Mechanistic and structural basis for activation of cardiac myosin force production by omecamtiv mecarbil.
Nat Commun 8(1):190 (2017)
Planelles-Herrero VJ, Hartman JJ, Robert-Paganin J, Malik FI, Houdusse A
|
| RgGuinier |
4.0 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
161 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Class1 collagenase monomer, 27 kDa Hathewaya histolytica protein
|
| Buffer: |
10 mM HEPES 100 mM NaCL, 2% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2015 Sep 14
|
Structural characterization of bacterial collagenases
University of Arkansas PhD thesis 2431 (2017)
Bauer R
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
36 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Class1 collagenase collagen-binding domain monomer, 27 kDa Hathewaya histolytica protein
|
| Buffer: |
10 mM HEPES 100 mM NaCL, 2% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2015 Sep 14
|
Structural characterization of bacterial collagenases
University of Arkansas PhD thesis 2431 (2017)
Bauer R
|
| RgGuinier |
2.4 |
nm |
| Dmax |
9.8 |
nm |
| VolumePorod |
33 |
nm3 |
|
|
|
|
|
|

|
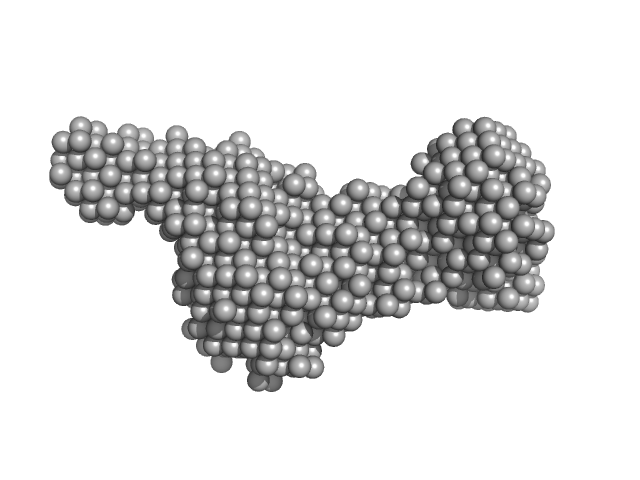
| Sample: |
Class1 collagenase monomer, 27 kDa Hathewaya histolytica protein
|
| Buffer: |
10 mM HEPES 100 mM NaCL, 2% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Sep 14
|
Structural characterization of bacterial collagenases
University of Arkansas PhD thesis 2431 (2017)
Bauer R
|
| RgGuinier |
2.5 |
nm |
| Dmax |
9.4 |
nm |
| VolumePorod |
36 |
nm3 |
|
|
|
|
|
|
|
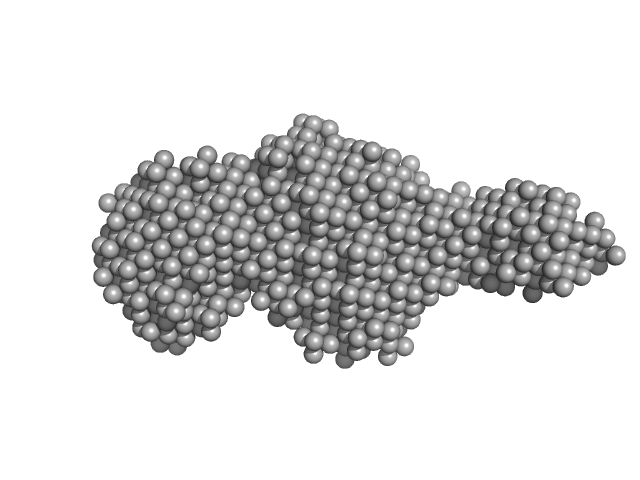
| Sample: |
Class1 collagenase monomer, 27 kDa Hathewaya histolytica protein
|
| Buffer: |
10 mM HEPES 100 mM NaCL, 2% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2015 Sep 14
|
Structural characterization of bacterial collagenases
University of Arkansas PhD thesis 2431 (2017)
Bauer R
|
| RgGuinier |
2.6 |
nm |
| Dmax |
12.8 |
nm |
| VolumePorod |
33 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nucleolysin TIA-1 isoform p40 monomer, 31 kDa Homo sapiens protein
|
| Buffer: |
10 mM Potassium Phosphate 50 mM NaCl 10 mM DTT, pH: 6 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, SFB 1035, Technische Universität München on 2015 Dec 1
|
Segmental, Domain-Selective Perdeuteration and Small-Angle Neutron Scattering for Structural Analysis of Multi-Domain Proteins.
Angew Chem Int Ed Engl 56(32):9322-9325 (2017)
Sonntag M, Jagtap PKA, Simon B, Appavou MS, Geerlof A, Stehle R, Gabel F, Hennig J, Sattler M
|
| RgGuinier |
2.7 |
nm |
| Dmax |
10.3 |
nm |
| VolumePorod |
38 |
nm3 |
|
|