|
|
|
|
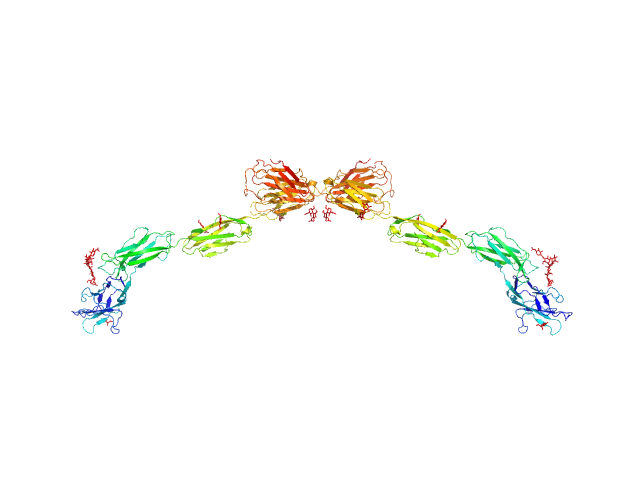
|
| Sample: |
Myelin-associated glycoprotein (20-508; N406Q mutant) monomer, 54 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Feb 5
|
Structural basis of myelin-associated glycoprotein adhesion and signalling.
Nat Commun 7:13584 (2016)
Pronker MF, Lemstra S, Snijder J, Heck AJ, Thies-Weesie DM, Pasterkamp RJ, Janssen BJ
|
| RgGuinier |
7.3 |
nm |
| Dmax |
25.6 |
nm |
| VolumePorod |
193 |
nm3 |
|
|
|
|
|
|
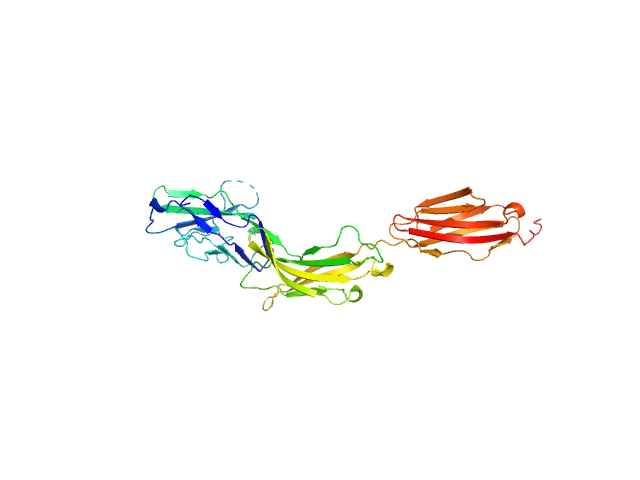
|
| Sample: |
Myelin-associated glycoprotein Ig domains 1-3 monomer, 35 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Sep 11
|
Structural basis of myelin-associated glycoprotein adhesion and signalling.
Nat Commun 7:13584 (2016)
Pronker MF, Lemstra S, Snijder J, Heck AJ, Thies-Weesie DM, Pasterkamp RJ, Janssen BJ
|
| RgGuinier |
3.9 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
59 |
nm3 |
|
|
|
|
|
|
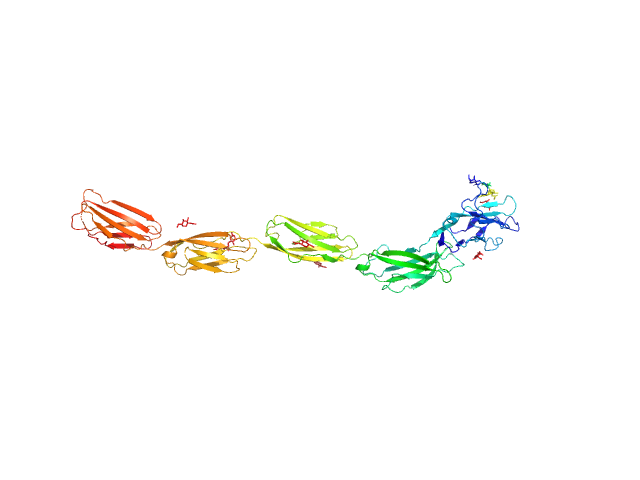
|
| Sample: |
Myelin-associated glycoprotein (20-508; I473E mutant) monomer, 54 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Sep 11
|
Structural basis of myelin-associated glycoprotein adhesion and signalling.
Nat Commun 7:13584 (2016)
Pronker MF, Lemstra S, Snijder J, Heck AJ, Thies-Weesie DM, Pasterkamp RJ, Janssen BJ
|
| RgGuinier |
6.0 |
nm |
| Dmax |
21.2 |
nm |
| VolumePorod |
100 |
nm3 |
|
|
|
|
|
|
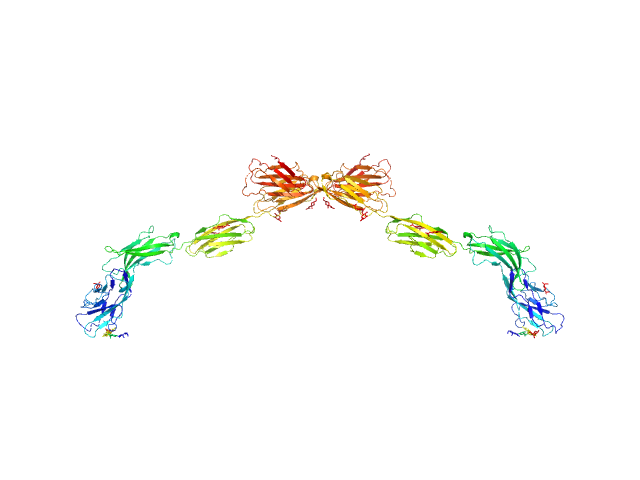
|
| Sample: |
Myelin-associated glycoprotein (20-508; N406Q mutant) monomer, 54 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Jul 28
|
Structural basis of myelin-associated glycoprotein adhesion and signalling.
Nat Commun 7:13584 (2016)
Pronker MF, Lemstra S, Snijder J, Heck AJ, Thies-Weesie DM, Pasterkamp RJ, Janssen BJ
|
| RgGuinier |
7.8 |
nm |
| Dmax |
29.0 |
nm |
| VolumePorod |
216 |
nm3 |
|
|
|
|
|
|
|
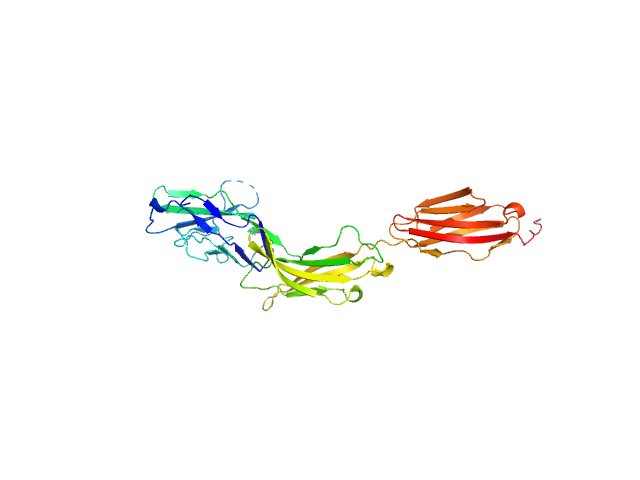
| Sample: |
Myelin-associated glycoprotein Ig domains 1-3 monomer, 35 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Sep 11
|
Structural basis of myelin-associated glycoprotein adhesion and signalling.
Nat Commun 7:13584 (2016)
Pronker MF, Lemstra S, Snijder J, Heck AJ, Thies-Weesie DM, Pasterkamp RJ, Janssen BJ
|
| RgGuinier |
3.9 |
nm |
| Dmax |
12.6 |
nm |
| VolumePorod |
49 |
nm3 |
|
|
|
|
|
|
|
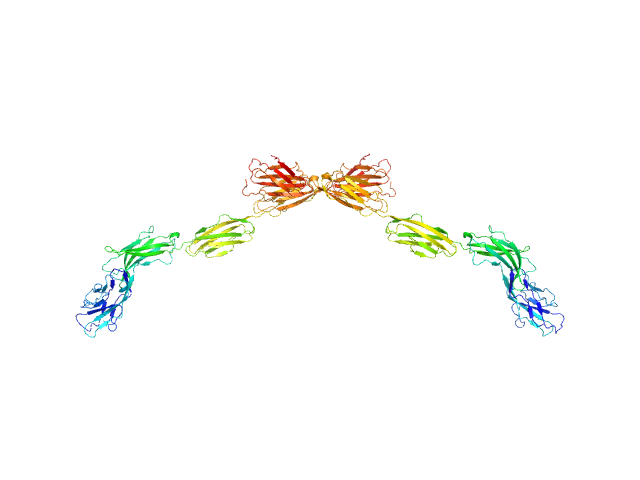
| Sample: |
Myelin-associated glycoprotein Ig domains 1-5 dimer, 108 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Sep 11
|
Structural basis of myelin-associated glycoprotein adhesion and signalling.
Nat Commun 7:13584 (2016)
Pronker MF, Lemstra S, Snijder J, Heck AJ, Thies-Weesie DM, Pasterkamp RJ, Janssen BJ
|
| RgGuinier |
7.3 |
nm |
| Dmax |
25.5 |
nm |
| VolumePorod |
166 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Cyclohexanone monooxygenase monomer, 61 kDa Rhodococcus sp. HI-31 protein
|
| Buffer: |
50 mM Tris, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2012 Oct 2
|
The role of conformational flexibility in Baeyer-Villiger monooxygenase catalysis and structure.
Biochim Biophys Acta 1864(12):1641-1648 (2016)
Yachnin BJ, Lau PCK, Berghuis AM
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.3 |
nm |
| VolumePorod |
110 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Cyclohexanone monooxygenase monomer, 61 kDa Rhodococcus sp. HI-31 protein
|
| Buffer: |
50 mM Tris 5 mM NADP+, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2013 Jan 27
|
The role of conformational flexibility in Baeyer-Villiger monooxygenase catalysis and structure.
Biochim Biophys Acta 1864(12):1641-1648 (2016)
Yachnin BJ, Lau PCK, Berghuis AM
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
99 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Cyclohexanone monooxygenase monomer, 61 kDa Rhodococcus sp. HI-31 protein
|
| Buffer: |
50 mM Tris 5 mM NADP+ 5 mM cyclohexanone, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2012 Oct 2
|
The role of conformational flexibility in Baeyer-Villiger monooxygenase catalysis and structure.
Biochim Biophys Acta 1864(12):1641-1648 (2016)
Yachnin BJ, Lau PCK, Berghuis AM
|
| RgGuinier |
2.5 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
100 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Cyclohexanone monooxygenase monomer, 61 kDa Rhodococcus sp. HI-31 protein
|
| Buffer: |
50 mM Tris 5 mM NADP+ 5 mM ε-caprolactone, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2012 Oct 2
|
The role of conformational flexibility in Baeyer-Villiger monooxygenase catalysis and structure.
Biochim Biophys Acta 1864(12):1641-1648 (2016)
Yachnin BJ, Lau PCK, Berghuis AM
|
| RgGuinier |
2.5 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
99 |
nm3 |
|
|