|
|
|
|
|
| Sample: |
Heterogeneous nuclear ribonucleoprotein A1 (C43S/R75D/R88D/C175S ) monomer, 25 kDa Homo sapiens protein
|
| Buffer: |
100 mM HEPES, 350 mM NaCl, 0.5 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2023 Mar 1
|
Thermodynamic coupling of the tandem RRM domains of hnRNP A1 underlie its pleiotropic RNA binding functions.
Sci Adv 10(28):eadk6580 (2024)
Levengood JD, Potoyan D, Penumutchu S, Kumar A, Zhou Q, Wang Y, Hansen AL, Kutluay S, Roche J, Tolbert BS
|
|
|
|
|
|
|
|
| Sample: |
Heterogeneous nuclear ribonucleoprotein A1 (C43S/R75D/R88D/C175S ) monomer, 25 kDa Homo sapiens protein
|
| Buffer: |
100 mM HEPES, 350 mM NaCl, 0.5 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2023 Mar 1
|
Thermodynamic coupling of the tandem RRM domains of hnRNP A1 underlie its pleiotropic RNA binding functions.
Sci Adv 10(28):eadk6580 (2024)
Levengood JD, Potoyan D, Penumutchu S, Kumar A, Zhou Q, Wang Y, Hansen AL, Kutluay S, Roche J, Tolbert BS
|
|
|
|
|
|
|
|
| Sample: |
Heterogeneous nuclear ribonucleoprotein A1 (C43S/R75D/R88D/C175S ) monomer, 25 kDa Homo sapiens protein
|
| Buffer: |
100 mM HEPES, 350 mM NaCl, 0.5 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2023 Mar 1
|
Thermodynamic coupling of the tandem RRM domains of hnRNP A1 underlie its pleiotropic RNA binding functions.
Sci Adv 10(28):eadk6580 (2024)
Levengood JD, Potoyan D, Penumutchu S, Kumar A, Zhou Q, Wang Y, Hansen AL, Kutluay S, Roche J, Tolbert BS
|
|
|
|
|
|
|
|
| Sample: |
Heterogeneous nuclear ribonucleoprotein A1 (C43S/R75D/R88D/C175S ) monomer, 25 kDa Homo sapiens protein
|
| Buffer: |
100 mM HEPES, 350 mM NaCl, 0.5 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2023 Mar 1
|
Thermodynamic coupling of the tandem RRM domains of hnRNP A1 underlie its pleiotropic RNA binding functions.
Sci Adv 10(28):eadk6580 (2024)
Levengood JD, Potoyan D, Penumutchu S, Kumar A, Zhou Q, Wang Y, Hansen AL, Kutluay S, Roche J, Tolbert BS
|
|
|
|
|
|
|
|
| Sample: |
Heterogeneous nuclear ribonucleoprotein A1 (C43S/R75D/R88D/C175S ) monomer, 25 kDa Homo sapiens protein
|
| Buffer: |
100 mM HEPES, 350 mM NaCl, 0.5 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2023 Mar 1
|
Thermodynamic coupling of the tandem RRM domains of hnRNP A1 underlie its pleiotropic RNA binding functions.
Sci Adv 10(28):eadk6580 (2024)
Levengood JD, Potoyan D, Penumutchu S, Kumar A, Zhou Q, Wang Y, Hansen AL, Kutluay S, Roche J, Tolbert BS
|
|
|
|
|
|
|
|
| Sample: |
Zinc finger and BTB domain-containing protein 8A.1-A dimer, 35 kDa Xenopus laevis protein
|
| Buffer: |
20 mM Hepes, 150 mM NaCl, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Dec 4
|
Dynamic BTB-domain filaments promote clustering of ZBTB proteins.
Mol Cell 84(13):2490-2510.e9 (2024)
Mance L, Bigot N, Zhamungui Sánchez E, Coste F, Martín-González N, Zentout S, Biliškov M, Pukało Z, Mishra A, Chapuis C, Arteni AA, Lateur A, Goffinont S, Gaudon V, Talhaoui I, Casuso I, Beaufour M, Garnier N, Artzner F, Cadene M, Huet S, Castaing B, Suskiewicz MJ
|
| RgGuinier |
22.3 |
nm |
| Dmax |
99.6 |
nm |
|
|
|
|
|
|
|
| Sample: |
Zinc finger and BTB domain-containing protein 8A.1-A (S103R mutant) dimer, 35 kDa Xenopus laevis protein
|
| Buffer: |
20 mM Hepes, 150 mM NaCl, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Dec 4
|
Dynamic BTB-domain filaments promote clustering of ZBTB proteins.
Mol Cell 84(13):2490-2510.e9 (2024)
Mance L, Bigot N, Zhamungui Sánchez E, Coste F, Martín-González N, Zentout S, Biliškov M, Pukało Z, Mishra A, Chapuis C, Arteni AA, Lateur A, Goffinont S, Gaudon V, Talhaoui I, Casuso I, Beaufour M, Garnier N, Artzner F, Cadene M, Huet S, Castaing B, Suskiewicz MJ
|
| RgGuinier |
3.3 |
nm |
| Dmax |
12.8 |
nm |
| VolumePorod |
60 |
nm3 |
|
|
|
|
|
|
|
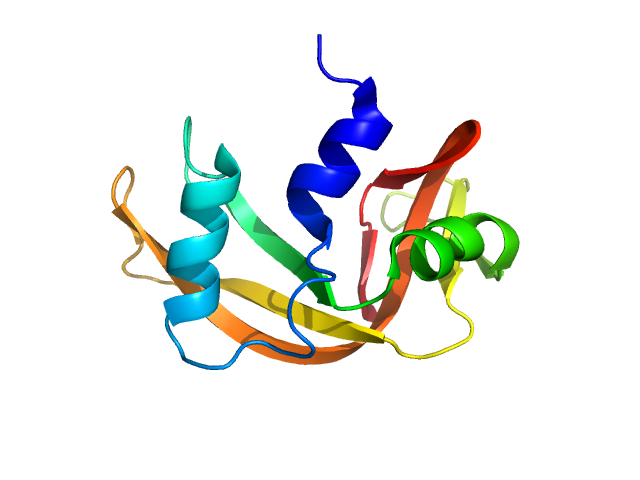
| Sample: |
Ribonuclease pancreatic monomer, 14 kDa Bos taurus protein
|
| Buffer: |
50 mM Tris, 100 mM NaCl, pH: 7.5 |
| Experiment: |
SANS
data collected at (Consensus SAS), Multi-facility, Multiple countries on 2024 Feb 4
|
Benchmarking predictive methods for small-angle X-ray scattering from atomic coordinates of proteins using maximum likelihood consensus data
IUCrJ 11(5) (2024)
Trewhella J, Vachette P, Larsen A
|
| RgGuinier |
1.5 |
nm |
| Dmax |
5.0 |
nm |
|
|
|
|
|
|
|
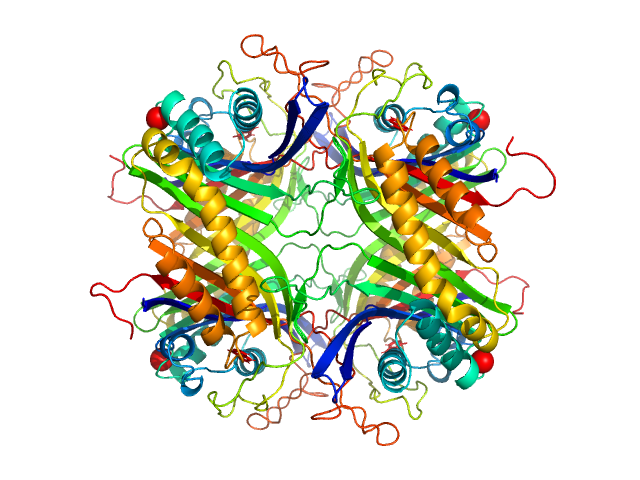
| Sample: |
Uricase tetramer, 136 kDa Aspergillus flavus protein
|
| Buffer: |
100 mM Tris, 150 mM NaCl, pH: 8 |
| Experiment: |
SANS
data collected at (Consensus SAS), Multi-facility, Multiple countries on 2024 Feb 4
|
Benchmarking predictive methods for small-angle X-ray scattering from atomic coordinates of proteins using maximum likelihood consensus data
IUCrJ 11(5) (2024)
Trewhella J, Vachette P, Larsen A
|
| RgGuinier |
3.2 |
nm |
| Dmax |
9.3 |
nm |
|
|
|
|
|
|
|
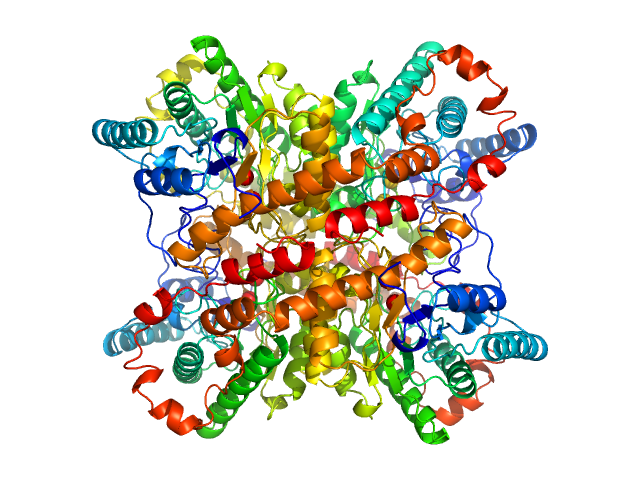
| Sample: |
Xylose isomerase tetramer, 173 kDa Streptomyces rubiginosus protein
|
| Buffer: |
50 mM Tris, 100 mM NaCl, 1 mM MgCl2, pH: 7.5 |
| Experiment: |
SANS
data collected at (Consensus SAS), Multi-facility, Multiple countries on 2024 Feb 4
|
Benchmarking predictive methods for small-angle X-ray scattering from atomic coordinates of proteins using maximum likelihood consensus data
IUCrJ 11(5) (2024)
Trewhella J, Vachette P, Larsen A
|
| RgGuinier |
3.3 |
nm |
| Dmax |
10.1 |
nm |
|
|