|
|
|
|
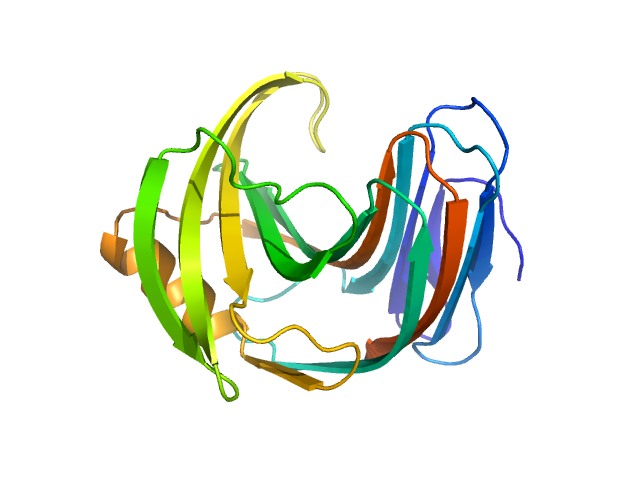
|
| Sample: |
Endo-1,4-beta-xylanase monomer, 21 kDa Trichoderma longibrachiatum protein
|
| Buffer: |
50 mM Tris, 100 mM NaCl, pH: 7.5 |
| Experiment: |
SANS
data collected at (Consensus SAS), Multi-facility, Multiple countries on 2024 Feb 4
|
Benchmarking predictive methods for small-angle X-ray scattering from atomic coordinates of proteins using maximum likelihood consensus data
IUCrJ 11(5) (2024)
Trewhella J, Vachette P, Larsen A
|
| RgGuinier |
1.6 |
nm |
| Dmax |
5.2 |
nm |
|
|
|
|
|
|
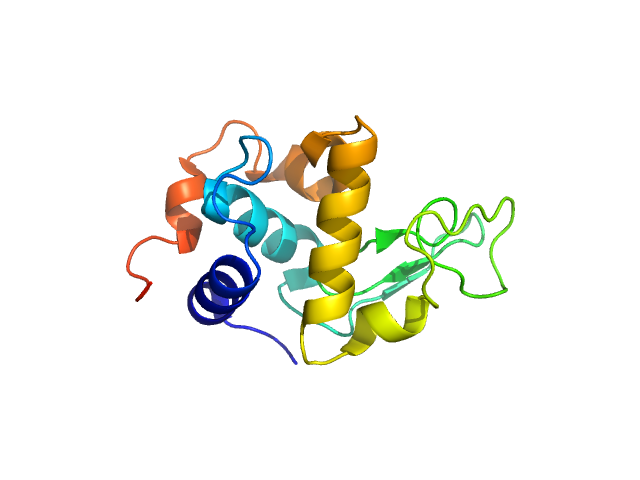
|
| Sample: |
Lysozyme C monomer, 14 kDa Gallus gallus protein
|
| Buffer: |
50 mM sodium citrate, 150 mM NaCl, pH: 4.5 |
| Experiment: |
SANS
data collected at (Consensus SAS), Multi-facility, Multiple countries on 2024 Feb 4
|
Benchmarking predictive methods for small-angle X-ray scattering from atomic coordinates of proteins using maximum likelihood consensus data
IUCrJ 11(5) (2024)
Trewhella J, Vachette P, Larsen A
|
| RgGuinier |
1.5 |
nm |
| Dmax |
4.7 |
nm |
|
|
|
|
|
|

|
| Sample: |
Z-DNA-binding protein 1 monomer, 18 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2018 Mar 23
|
ZBP1 condensate formation synergizes Z-NAs recognition and signal transduction.
Cell Death Dis 15(7):487 (2024)
Xie F, Wu D, Huang J, Liu X, Shen Y, Huang J, Su Z, Li J
|
| RgGuinier |
2.1 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
19 |
nm3 |
|
|
|
|
|
|
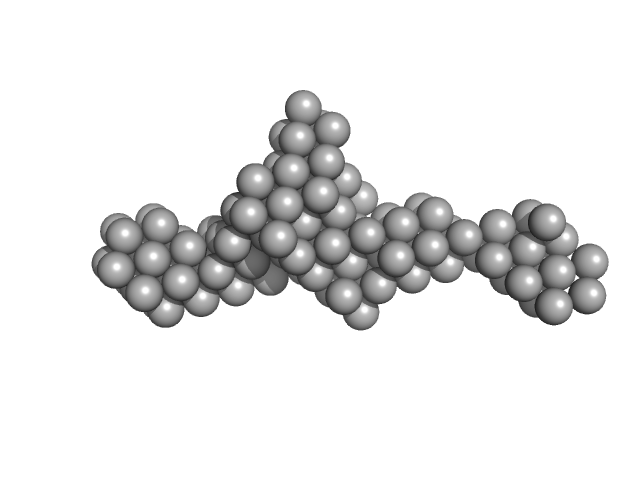
|
| Sample: |
Z-DNA-binding protein 1 dimer, 37 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2018 Mar 23
|
ZBP1 condensate formation synergizes Z-NAs recognition and signal transduction.
Cell Death Dis 15(7):487 (2024)
Xie F, Wu D, Huang J, Liu X, Shen Y, Huang J, Su Z, Li J
|
| RgGuinier |
3.1 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
48 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Iron-sulfur cluster assembly 1 homolog, mitochondrial, 15 kDa Columba livia protein
|
| Buffer: |
20 mM Tris-HCl, 0.15 M NaCl, 10 mM 3-mercapto-1,2-propanediol, pH: 8 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2020 Nov 30
|
Radiation resistivity of clISCA1
Shigeki Arai
|
| RgGuinier |
4.3 |
nm |
| Dmax |
15.8 |
nm |
|
|
|
|
|
|
|
| Sample: |
Pigeon iron-sulfur cluster assembly 1 homolog, mitochondrial, 15 kDa Columba livia protein
|
| Buffer: |
20 mM Tris-HCl, 0.15 M NaCl, 10 mM 3-mercapto-1,2-propanediol, pH: 8 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2020 Nov 30
|
Radiation resistivity of clISCA1
Shigeki Arai
|
|
|
|
|
|
|
|
| Sample: |
Iron-sulfur cluster assembly 1 homolog, mitochondrial, 15 kDa Columba livia protein
|
| Buffer: |
20 mM Tris-HCl, 0.15 M NaCl, 10 mM 3-mercapto-1,2-propanediol, pH: 8 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2020 Nov 30
|
Radiation resistivity of clISCA1
Shigeki Arai
|
|
|
|
|
|
|
|
| Sample: |
Iron-sulfur cluster assembly 1 homolog, mitochondrial, 15 kDa Columba livia protein
|
| Buffer: |
20 mM Tris-HCl, 0.15 M NaCl, 10 mM 3-mercapto-1,2-propanediol, pH: 8 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2020 Nov 30
|
Radiation resistivity of clISCA1
Shigeki Arai
|
|
|
|
|
|
|
|
| Sample: |
Iron-sulfur cluster assembly 1 homolog, mitochondrial, 15 kDa Columba livia protein
|
| Buffer: |
20 mM Tris-HCl, 0.15 M NaCl, 10 mM 3-mercapto-1,2-propanediol, pH: 8 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2020 Nov 30
|
Radiation resistivity of clISCA1
Shigeki Arai
|
|
|
|
|
|
|
|
| Sample: |
Iron-sulfur cluster assembly 1 homolog, mitochondrial, 15 kDa Columba livia protein
|
| Buffer: |
20 mM Tris-HCl, 0.15 M NaCl, 10 mM 3-mercapto-1,2-propanediol, pH: 8 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2021 Jun 8
|
Radiation resistivity of clISCA1
Shigeki Arai
|
|
|