|
|
|
|
|
| Sample: |
Lysozyme C monomer, 14 kDa Gallus gallus protein
|
| Buffer: |
1 mol% ethylammonium nitrate, pH: 8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Nov 27
|
Scattering approaches to unravel protein solution behaviors in ionic liquids and deep eutectic solvents: From basic principles to recent developments
Advances in Colloid and Interface Science :103242 (2024)
Han Q, Veríssimo N, Bryant S, Martin A, Huang Y, Pereira J, Santos-Ebinuma V, Zhai J, Bryant G, Drummond C, Greaves T
|
| RgGuinier |
1.6 |
nm |
| Dmax |
6.2 |
nm |
| VolumePorod |
28 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Lysozyme C monomer, 14 kDa Gallus gallus protein
|
| Buffer: |
1 mol% ethylammonium nitrate, pH: 8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Nov 27
|
Scattering approaches to unravel protein solution behaviors in ionic liquids and deep eutectic solvents: From basic principles to recent developments
Advances in Colloid and Interface Science :103242 (2024)
Han Q, Veríssimo N, Bryant S, Martin A, Huang Y, Pereira J, Santos-Ebinuma V, Zhai J, Bryant G, Drummond C, Greaves T
|
| RgGuinier |
1.6 |
nm |
| Dmax |
6.2 |
nm |
| VolumePorod |
23 |
nm3 |
|
|
|
|
|
|
|
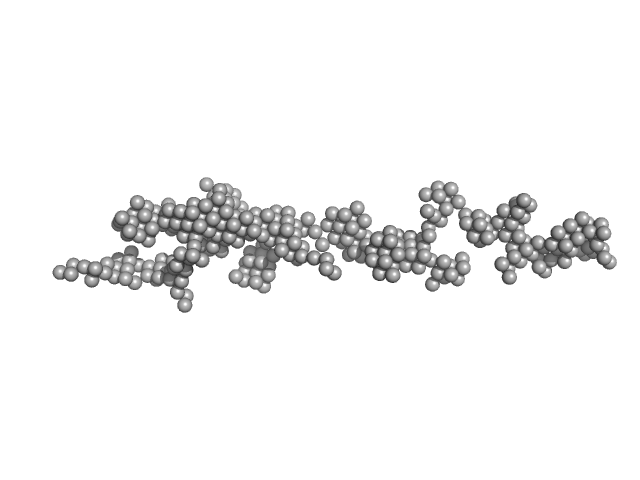
| Sample: |
DNA polymerase monomer, 117 kDa Vaccinia virus (strain … protein
DNA polymerase processivity factor component OPG148 monomer, 49 kDa Vaccinia virus (strain … protein
Uracil-DNA glycosylase monomer, 27 kDa Vaccinia virus (strain … protein
|
| Buffer: |
200 mM NaCl, 20 mM Tris-HCl and 1 mM TCEP, pH: 8.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Jan 25
|
Structure and flexibility of the DNA polymerase holoenzyme of vaccinia virus.
PLoS Pathog 20(5):e1011652 (2024)
Burmeister WP, Boutin L, Balestra AC, Gröger H, Ballandras-Colas A, Hutin S, Kraft C, Grimm C, Böttcher B, Fischer U, Tarbouriech N, Iseni F
|
| RgGuinier |
5.0 |
nm |
| Dmax |
18.0 |
nm |
| VolumePorod |
350 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Amyloid-beta precursor protein, 5 kDa Homo sapiens protein
|
| Buffer: |
1 mM Hepes, 0.12 % NH4OH, pH: 10.7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Apr 11
|
Time-resolved small-angle X-ray scattering study of polymorphic nature of Aß42 oligomerisation
Tia Cheremnykh
|
| RgGuinier |
5.8 |
nm |
| Dmax |
42.0 |
nm |
|
|
|
|
|
|

|
| Sample: |
Amyloid-beta precursor protein, 5 kDa Homo sapiens protein
|
| Buffer: |
1 mM Hepes, 0.12 % NH4OH, pH: 10.7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Dec 13
|
Time-resolved small-angle X-ray scattering study of polymorphic nature of Aß42 oligomerisation
Tia Cheremnykh
|
| RgGuinier |
6.9 |
nm |
| Dmax |
60.0 |
nm |
|
|
|
|
|
|

|
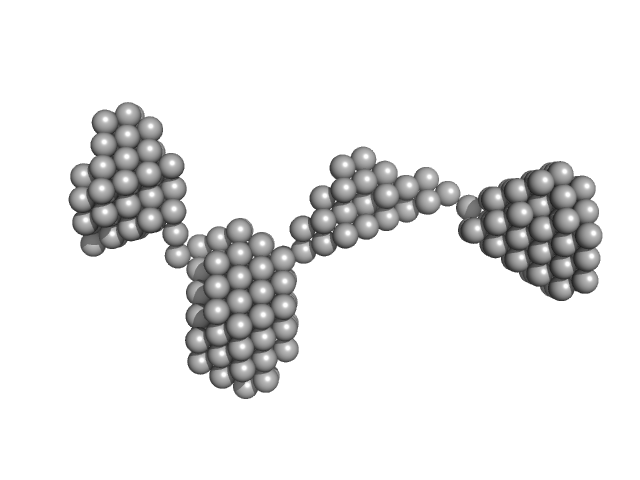
| Sample: |
Amyloid-beta precursor protein, 5 kDa Homo sapiens protein
|
| Buffer: |
1 mM Hepes, 0.1 % NH4OH (pH∼10.7), pH: 10.7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Oct 31
|
Time-resolved small-angle X-ray scattering study of polymorphic nature of Aß42 oligomerisation
Tia Cheremnykh
|
| RgGuinier |
2.5 |
nm |
| Dmax |
6.5 |
nm |
|
|
|
|
|
|
|
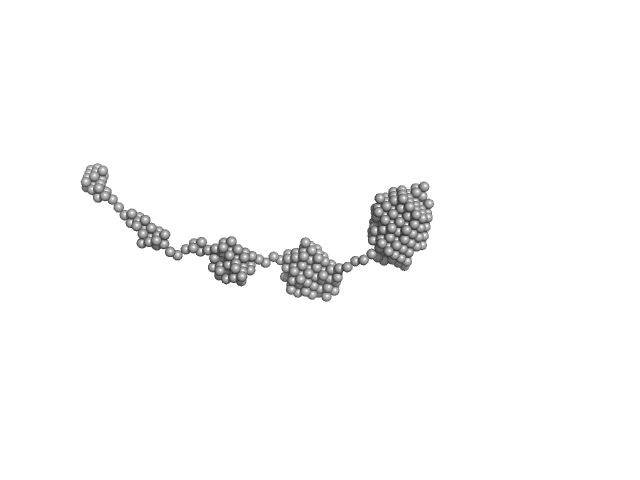
| Sample: |
Amyloid-beta precursor protein, 5 kDa Homo sapiens protein
|
| Buffer: |
1 mM Hepes, 0.1 % NH4OH (pH∼10.7), pH: 10.7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Nov 1
|
Time-resolved small-angle X-ray scattering study of polymorphic nature of Aß42 oligomerisation
Tia Cheremnykh
|
| RgGuinier |
3.7 |
nm |
| Dmax |
9.1 |
nm |
|
|
|
|
|
|
|
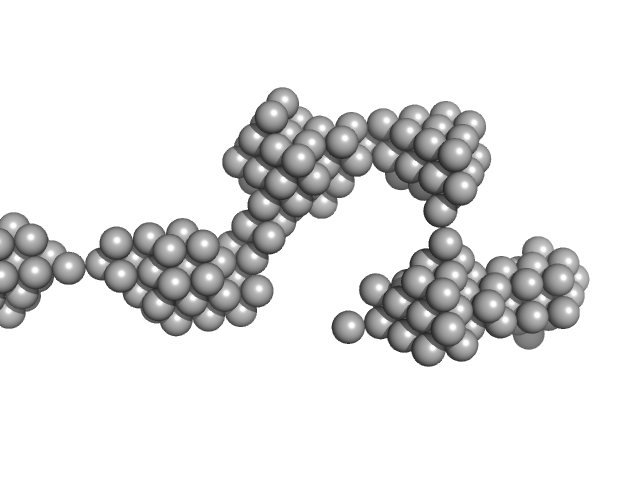
| Sample: |
Amyloid-beta precursor protein, 5 kDa Homo sapiens protein
|
| Buffer: |
1 mM Hepes, 0.1 % NH4OH (pH∼10.7), pH: 10.7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Nov 1
|
Time-resolved small-angle X-ray scattering study of polymorphic nature of Aß42 oligomerisation
Tia Cheremnykh
|
| RgGuinier |
3.7 |
nm |
| Dmax |
11.0 |
nm |
|
|
|
|
|
|
|
| Sample: |
Amyloid-beta precursor protein, 5 kDa Homo sapiens protein
|
| Buffer: |
1 mM Hepes, 0.12 % NH4OH, pH: 10.7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Nov 2
|
Time-resolved small-angle X-ray scattering study of polymorphic nature of Aß42 oligomerisation
Tia Cheremnykh
|
| RgGuinier |
3.5 |
nm |
| Dmax |
15.0 |
nm |
|
|
|
|
|
|
|
| Sample: |
Mucin-2 (CysD2 domain) monomer, 16 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 70 mM NaCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Mar 8
|
The structure of the second CysD domain of MUC2 and role in mucin organization by transglutaminase-based cross-linking.
Cell Rep 43(5):114207 (2024)
Recktenwald CV, Karlsson G, Garcia-Bonete MJ, Katona G, Jensen M, Lymer R, Bäckström M, Johansson MEV, Hansson GC, Trillo-Muyo S
|
| RgGuinier |
2.5 |
nm |
| Dmax |
9.3 |
nm |
| VolumePorod |
31 |
nm3 |
|
|