|
|
|
|
|
| Sample: |
29 kDa ribonucleoprotein, chloroplastic monomer, 8 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM sodium phosphate, 30 mM NaCl, pH: 6.2 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, SFB 1035, Technische Universität München on 2021 Oct 19
|
A prion-like domain is required for phase separation and chloroplast RNA processing during cold acclimation in Arabidopsis.
Plant Cell (2024)
Legen J, Lenzen B, Kachariya N, Feltgen S, Gao Y, Mergenthal S, Weber W, Klotzsch E, Zoschke R, Sattler M, Schmitz-Linneweber C
|
| RgGuinier |
2.1 |
nm |
| Dmax |
9.2 |
nm |
| VolumePorod |
19 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
29 kDa ribonucleoprotein, chloroplastic monomer, 8 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM sodium phosphate, 30 mM NaCl, pH: 6.2 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, SFB 1035, Technische Universität München on 2021 Oct 19
|
A prion-like domain is required for phase separation and chloroplast RNA processing during cold acclimation in Arabidopsis.
Plant Cell (2024)
Legen J, Lenzen B, Kachariya N, Feltgen S, Gao Y, Mergenthal S, Weber W, Klotzsch E, Zoschke R, Sattler M, Schmitz-Linneweber C
|
| RgGuinier |
2.4 |
nm |
| Dmax |
11.6 |
nm |
| VolumePorod |
25 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Pre-mRNA-processing factor 40 homolog A monomer, 11 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 100 mM NaCl, 0.5 mM DTT, pH: 6.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2020 Sep 19
|
Intramolecular autoinhibition regulates the selectivity of PRPF40A tandem WW domains for proline-rich motifs.
Nat Commun 15(1):3888 (2024)
Martínez-Lumbreras S, Träger LK, Mulorz MM, Payr M, Dikaya V, Hipp C, König J, Sattler M
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.4 |
nm |
| VolumePorod |
17 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Pre-mRNA-processing factor 40 homolog A monomer, 11 kDa Homo sapiens protein
Splicing factor 1 monomer, 2 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 100 mM NaCl, 0.5 mM DTT, pH: 6.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2020 Sep 19
|
Intramolecular autoinhibition regulates the selectivity of PRPF40A tandem WW domains for proline-rich motifs.
Nat Commun 15(1):3888 (2024)
Martínez-Lumbreras S, Träger LK, Mulorz MM, Payr M, Dikaya V, Hipp C, König J, Sattler M
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.9 |
nm |
| VolumePorod |
19 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Pre-mRNA-processing factor 40 homolog A monomer, 11 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 1 mM DTT, pH: 6.6 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, Technische Universität München on 2024 Mar 8
|
Intramolecular autoinhibition regulates the selectivity of PRPF40A tandem WW domains for proline-rich motifs.
Nat Commun 15(1):3888 (2024)
Martínez-Lumbreras S, Träger LK, Mulorz MM, Payr M, Dikaya V, Hipp C, König J, Sattler M
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.3 |
nm |
| VolumePorod |
18 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Pre-mRNA-processing factor 40 homolog A monomer, 11 kDa Homo sapiens protein
Splicing factor 1 monomer, 2 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 1 mM DTT, pH: 6.6 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, Technische Universität München on 2024 Mar 8
|
Intramolecular autoinhibition regulates the selectivity of PRPF40A tandem WW domains for proline-rich motifs.
Nat Commun 15(1):3888 (2024)
Martínez-Lumbreras S, Träger LK, Mulorz MM, Payr M, Dikaya V, Hipp C, König J, Sattler M
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.0 |
nm |
| VolumePorod |
19 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Endonuclease 8 2 dimer, 108 kDa Mycobacterium tuberculosis (strain … protein
Single-stranded DNA-binding protein octamer, 139 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR-Central Drug Research Institute on 2019 Jul 10
|
Identification and characterization of MtbNei2-SSB forms novel pre-replicative BER complex in Mycobacterium tuberculosis.
Jyoti Vishwakarma
|
| RgGuinier |
5.5 |
nm |
| Dmax |
21.0 |
nm |
| VolumePorod |
734 |
nm3 |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDQZ7_fit2_model1.png)
|
| Sample: |
Ubiquitin carboxyl-terminal hydrolase 14 monomer, 56 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Oct 18
|
Transient interdomain interactions in free USP14 shape its conformational ensemble.
Protein Sci 33(5):e4975 (2024)
Salomonsson J, Wallner B, Sjöstrand L, D'Arcy P, Sunnerhagen M, Ahlner A
|
| RgGuinier |
3.1 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
111 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Ubiquitin carboxyl-terminal hydrolase 14 monomer, 46 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jul 5
|
Transient interdomain interactions in free USP14 shape its conformational ensemble.
Protein Sci 33(5):e4975 (2024)
Salomonsson J, Wallner B, Sjöstrand L, D'Arcy P, Sunnerhagen M, Ahlner A
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.3 |
nm |
| VolumePorod |
88 |
nm3 |
|
|
|
|
|
|
|
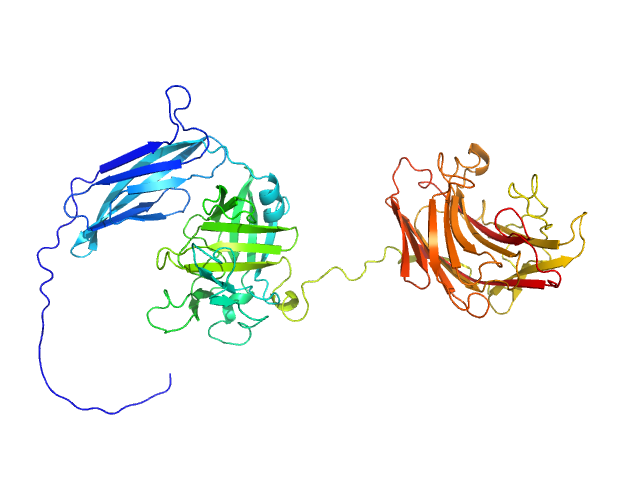
| Sample: |
Glycosyl hydrolase, family 16 monomer, 58 kDa Christiangramia forsetii (strain … protein
|
| Buffer: |
10 mM MOPS, 100 mM NaCl, pH: 7.8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Dec 1
|
Unveiling the role of novel carbohydrate-binding modules in laminarin interaction of multimodular proteins from marine Bacteroidota during phytoplankton blooms.
Environ Microbiol 26(5):e16624 (2024)
Zühlke MK, Ficko-Blean E, Bartosik D, Terrapon N, Jeudy A, Jam M, Wang F, Welsch N, Dürwald A, Martin LT, Larocque R, Jouanneau D, Eisenack T, Thomas F, Trautwein-Schult A, Teeling H, Becher D, Schweder T, Czjzek M
|
| RgGuinier |
3.8 |
nm |
| Dmax |
12.9 |
nm |
| VolumePorod |
76 |
nm3 |
|
|