|
|
|
|

|
| Sample: |
Beta-glucosidase dimer, 187 kDa Aspergillus niger protein
|
| Buffer: |
50 mM sodium acetate, pH: 5 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2024 Feb 15
|
Aspergillus niger beta-glucosidase (bgl1)
Estella Yee
|
| RgGuinier |
4.2 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
253 |
nm3 |
|
|
|
|
|
|
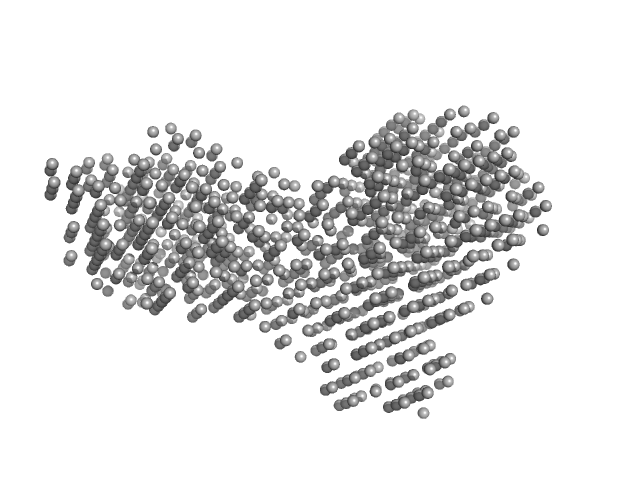
|
| Sample: |
Beta-glucosidase dimer, 187 kDa Aspergillus niger protein
|
| Buffer: |
50 mM sodium acetate, pH: 5 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2024 Jun 4
|
Aspergillus niger beta-glucosidase (bgl1)
Estella Yee
|
| RgGuinier |
4.4 |
nm |
| Dmax |
13.9 |
nm |
| VolumePorod |
277 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Isoform A1B1 of Teneurin-3 dimer, 545 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Sep 10
|
Alternative splicing controls teneurin-3 compact dimer formation for neuronal recognition
Nature Communications 15(1) (2024)
Gogou C, Beugelink J, Frias C, Kresik L, Jaroszynska N, Drescher U, Janssen B, Hindges R, Meijer D
|
| RgGuinier |
8.9 |
nm |
| Dmax |
37.0 |
nm |
| VolumePorod |
1059 |
nm3 |
|
|
|
|
|
|
|
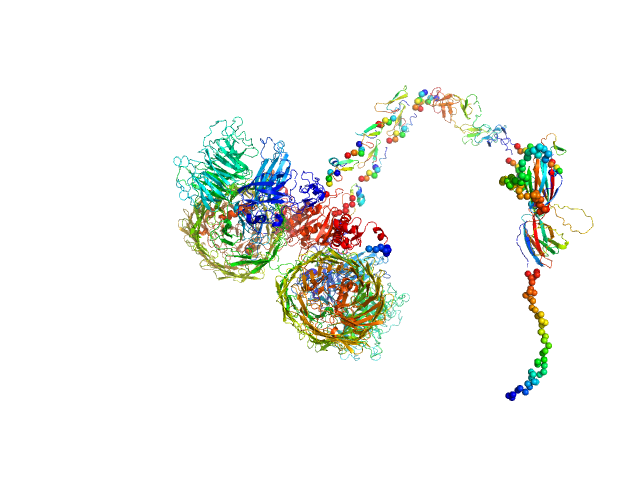
| Sample: |
Isoform A1B1 of Teneurin-3 dimer, 545 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2 mM CaCl2, pH: 7.8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Sep 10
|
Alternative splicing controls teneurin-3 compact dimer formation for neuronal recognition
Nature Communications 15(1) (2024)
Gogou C, Beugelink J, Frias C, Kresik L, Jaroszynska N, Drescher U, Janssen B, Hindges R, Meijer D
|
| RgGuinier |
7.9 |
nm |
| Dmax |
33.0 |
nm |
| VolumePorod |
1133 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Isoform A1B1 of Teneurin-3 dimer, 545 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2 mM CaCl2, pH: 7.8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Sep 10
|
Alternative splicing controls teneurin-3 compact dimer formation for neuronal recognition
Nature Communications 15(1) (2024)
Gogou C, Beugelink J, Frias C, Kresik L, Jaroszynska N, Drescher U, Janssen B, Hindges R, Meijer D
|
| RgGuinier |
7.9 |
nm |
| Dmax |
32.0 |
nm |
| VolumePorod |
1129 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Isoform A1B1 of Teneurin-3 dimer, 545 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2 mM CaCl2, pH: 7.8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Sep 10
|
Alternative splicing controls teneurin-3 compact dimer formation for neuronal recognition
Nature Communications 15(1) (2024)
Gogou C, Beugelink J, Frias C, Kresik L, Jaroszynska N, Drescher U, Janssen B, Hindges R, Meijer D
|
| RgGuinier |
7.9 |
nm |
| Dmax |
33.0 |
nm |
| VolumePorod |
1129 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Isoform A1B1 of Teneurin-3 dimer, 545 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 5 mM EDTA, pH: 7.8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Sep 10
|
Alternative splicing controls teneurin-3 compact dimer formation for neuronal recognition
Nature Communications 15(1) (2024)
Gogou C, Beugelink J, Frias C, Kresik L, Jaroszynska N, Drescher U, Janssen B, Hindges R, Meijer D
|
| RgGuinier |
8.8 |
nm |
| Dmax |
35.0 |
nm |
| VolumePorod |
1035 |
nm3 |
|
|
|
|
|
|
|
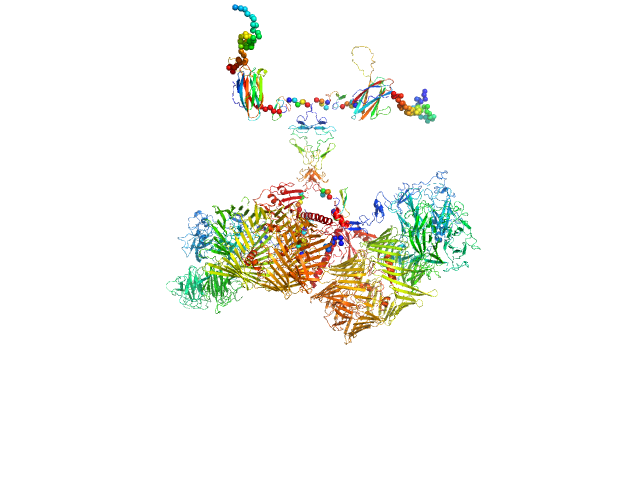
| Sample: |
Isoform A0B0 of Teneurin-3 dimer, 526 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Sep 10
|
Alternative splicing controls teneurin-3 compact dimer formation for neuronal recognition
Nature Communications 15(1) (2024)
Gogou C, Beugelink J, Frias C, Kresik L, Jaroszynska N, Drescher U, Janssen B, Hindges R, Meijer D
|
| RgGuinier |
9.4 |
nm |
| Dmax |
39.0 |
nm |
| VolumePorod |
1101 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Isoform A0B0 of Teneurin-3 dimer, 526 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2 mM CaCl2, pH: 7.8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Sep 10
|
Alternative splicing controls teneurin-3 compact dimer formation for neuronal recognition
Nature Communications 15(1) (2024)
Gogou C, Beugelink J, Frias C, Kresik L, Jaroszynska N, Drescher U, Janssen B, Hindges R, Meijer D
|
| RgGuinier |
7.3 |
nm |
| Dmax |
31.0 |
nm |
| VolumePorod |
1057 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Isoform A0B0 of Teneurin-3 dimer, 526 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2 mM CaCl2, pH: 7.8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Sep 10
|
Alternative splicing controls teneurin-3 compact dimer formation for neuronal recognition
Nature Communications 15(1) (2024)
Gogou C, Beugelink J, Frias C, Kresik L, Jaroszynska N, Drescher U, Janssen B, Hindges R, Meijer D
|
| RgGuinier |
7.3 |
nm |
| Dmax |
31.0 |
nm |
| VolumePorod |
1077 |
nm3 |
|
|