|
|
|
|
|
| Sample: |
Annexin A11 monomer, 54 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM KCl, 500 µM CaCl2, 1 mM DTT, pH: 8.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 Jul 21
|
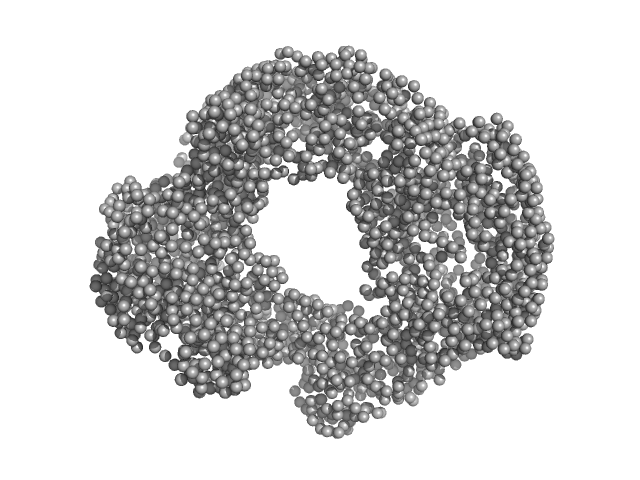
The structural properties of full-length annexin A11
Frontiers in Molecular Biosciences 11 (2024)
Dudas E, Tully M, Foldes T, Kelly G, Tartaglia G, Pastore A
|
| RgGuinier |
3.8 |
nm |
| Dmax |
17.1 |
nm |
| VolumePorod |
83 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Acylamino-acid-releasing enzyme (I277L, V491A) tetramer, 308 kDa Geobacillus stearothermophilus protein
|
| Buffer: |
10 mM Tris, 100 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL-18, INDUS-2 on 2023 Aug 11
|
Crystal structure and solution scattering of Geobacillus stearothermophilus S9 peptidase reveal structural adaptations for carboxypeptidase activity.
FEBS Lett (2024)
Chandravanshi K, Singh R, Bhange GN, Kumar A, Yadav P, Kumar A, Makde RD
|
| RgGuinier |
5.3 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
424 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Cadherin-2 dimer, 123 kDa Mus musculus protein
|
| Buffer: |
10 mM HEPES, 150 mM NaCl, 3 mM CaCl2, 0.02% NaN3, pH: 8 |
| Experiment: |
SAXS
data collected at 13A, Taiwan Photon Source, NSRRC on 2023 May 1
|
Rapid simulation of glycoprotein structures by grafting and steric exclusion of glycan conformer libraries.
Cell 187(5):1296-1311.e26 (2024)
Tsai YX, Chang NE, Reuter K, Chang HT, Yang TJ, von Bülow S, Sehrawat V, Zerrouki N, Tuffery M, Gecht M, Grothaus IL, Colombi Ciacchi L, Wang YS, Hsu MF, Khoo KH, Hummer G, Hsu SD, Hanus C, Sikora M
|
| RgGuinier |
8.8 |
nm |
| Dmax |
40.0 |
nm |
| VolumePorod |
496 |
nm3 |
|
|
|
|
|
|
|
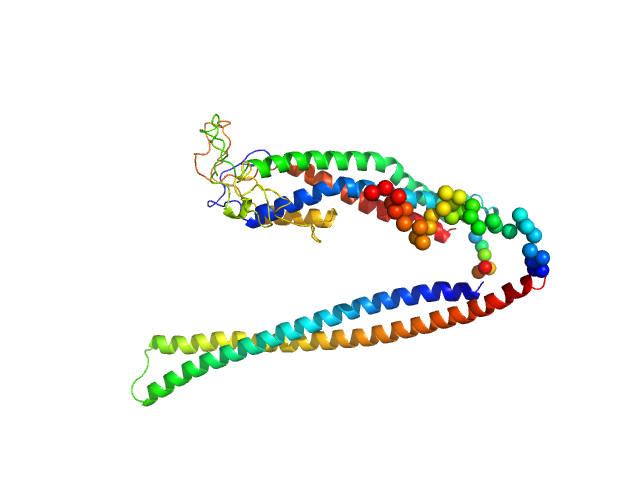
| Sample: |
Cadherin-2 monomer, 24 kDa Mus musculus protein
|
| Buffer: |
10 mM HEPES, 150 mM NaCl, 3 mM CaCl2, 0.02% NaN3, pH: 8 |
| Experiment: |
SAXS
data collected at 13A, Taiwan Photon Source, NSRRC on 2023 May 1
|
Rapid simulation of glycoprotein structures by grafting and steric exclusion of glycan conformer libraries.
Cell 187(5):1296-1311.e26 (2024)
Tsai YX, Chang NE, Reuter K, Chang HT, Yang TJ, von Bülow S, Sehrawat V, Zerrouki N, Tuffery M, Gecht M, Grothaus IL, Colombi Ciacchi L, Wang YS, Hsu MF, Khoo KH, Hummer G, Hsu SD, Hanus C, Sikora M
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.6 |
nm |
| VolumePorod |
63 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Invariant surface glycoprotein (N134A) monomer, 49 kDa Trypanosoma brucei protein
|
| Buffer: |
20 mM Tris-HCl, 75 mM KCl, pH: 7.6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Aug 18
|
Trypanosoma brucei Invariant Surface Glycoprotein 75 Is an Immunoglobulin Fc Receptor Inhibiting Complement Activation and Antibody-Mediated Cellular Phagocytosis.
J Immunol (2024)
Mikkelsen JH, Stødkilde K, Jensen MP, Hansen AG, Wu Q, Lorentzen J, Graversen JH, Andersen GR, Fenton RA, Etzerodt A, Thiel S, Andersen CBF
|
| RgGuinier |
3.6 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
127 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Netrin unc-6 monomer, 52 kDa Caenorhabditis elegans protein
Isoform a of Netrin receptor unc-5 monomer, 38 kDa Caenorhabditis elegans protein
Heparin, porcine intestinal mucosa monomer, 15 kDa Sus scrofa domesticus
|
| Buffer: |
10 mM HEPES pH 7.2, 150 mM NaCl, 100 mM MgSO4, pH: 7.2 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Jun 18
|
Structural insights into the formation of repulsive netrin guidance complexes.
Sci Adv 10(7):eadj8083 (2024)
Priest JM, Nichols EL, Smock RG, Hopkins JB, Mendoza JL, Meijers R, Shen K, Özkan E
|
| RgGuinier |
10.4 |
nm |
| Dmax |
37.3 |
nm |
| VolumePorod |
2156 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Netrin unc-6 monomer, 52 kDa Caenorhabditis elegans protein
Isoform a of Netrin receptor unc-5 monomer, 38 kDa Caenorhabditis elegans protein
Heparin, porcine intestinal mucosa monomer, 15 kDa Sus scrofa domesticus
Netrin receptor unc-40 monomer, 118 kDa Caenorhabditis elegans protein
|
| Buffer: |
10 mM HEPES pH 7.2, 150 mM NaCl, 100 mM MgSO4, pH: 7.2 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Dec 11
|
Structural insights into the formation of repulsive netrin guidance complexes.
Sci Adv 10(7):eadj8083 (2024)
Priest JM, Nichols EL, Smock RG, Hopkins JB, Mendoza JL, Meijers R, Shen K, Özkan E
|
| RgGuinier |
10.6 |
nm |
| Dmax |
36.5 |
nm |
| VolumePorod |
3410 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Neurofilament light polypeptide (T445N; C-terminus, amino acids 399-553), 16 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Sep 7
|
From isolated polyelectrolytes to star-like assemblies: the role of sequence heterogeneity on the statistical structure of the intrinsically disordered neurofilament-low tail domain.
Eur Phys J E Soft Matter 47(2):13 (2024)
Kravikass M, Koren G, Saleh OA, Beck R
|
|
|
|
|
|
|
|
| Sample: |
Neurofilament light polypeptide (T445N; C-terminus, amino acids 399-553), 16 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Sep 7
|
From isolated polyelectrolytes to star-like assemblies: the role of sequence heterogeneity on the statistical structure of the intrinsically disordered neurofilament-low tail domain.
Eur Phys J E Soft Matter 47(2):13 (2024)
Kravikass M, Koren G, Saleh OA, Beck R
|
|
|
|
|
|
|
|
| Sample: |
Neurofilament light polypeptide (T445N; C-terminus, amino acids 399-553), 16 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Sep 7
|
From isolated polyelectrolytes to star-like assemblies: the role of sequence heterogeneity on the statistical structure of the intrinsically disordered neurofilament-low tail domain.
Eur Phys J E Soft Matter 47(2):13 (2024)
Kravikass M, Koren G, Saleh OA, Beck R
|
|
|