|
|
|
|
|
| Sample: |
Glycosyl hydrolase family 61 monomer, 30 kDa Colletotrichum higginsianum (strain … protein
|
| Buffer: |
50 mM MES, 150 mM NaCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Jun 9
|
The disordered C-terminal tail of fungal LPMOs from phytopathogens mediates protein dimerization and impacts plant penetration.
Proc Natl Acad Sci U S A 121(13):e2319998121 (2024)
Tamburrini KC, Kodama S, Grisel S, Haon M, Nishiuchi T, Bissaro B, Kubo Y, Longhi S, Berrin JG
|
| RgGuinier |
4.1 |
nm |
| Dmax |
14.6 |
nm |
| VolumePorod |
85 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Glycosyl hydrolase family 61 dimer, 61 kDa Colletotrichum higginsianum (strain … protein
|
| Buffer: |
50 mM MES, 150 mM NaCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Jun 9
|
The disordered C-terminal tail of fungal LPMOs from phytopathogens mediates protein dimerization and impacts plant penetration.
Proc Natl Acad Sci U S A 121(13):e2319998121 (2024)
Tamburrini KC, Kodama S, Grisel S, Haon M, Nishiuchi T, Bissaro B, Kubo Y, Longhi S, Berrin JG
|
| RgGuinier |
6.2 |
nm |
| Dmax |
24.5 |
nm |
| VolumePorod |
206 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Endoglucanase-4 monomer, 33 kDa Colletotrichum higginsianum (strain … protein
|
| Buffer: |
50 mM MES, 150 mM NaCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Feb 17
|
The disordered C-terminal tail of fungal LPMOs from phytopathogens mediates protein dimerization and impacts plant penetration.
Proc Natl Acad Sci U S A 121(13):e2319998121 (2024)
Tamburrini KC, Kodama S, Grisel S, Haon M, Nishiuchi T, Bissaro B, Kubo Y, Longhi S, Berrin JG
|
| RgGuinier |
3.9 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
68 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Endoglucanase-4 dimer, 66 kDa Colletotrichum higginsianum (strain … protein
|
| Buffer: |
50 mM MES, 150 mM NaCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Feb 17
|
The disordered C-terminal tail of fungal LPMOs from phytopathogens mediates protein dimerization and impacts plant penetration.
Proc Natl Acad Sci U S A 121(13):e2319998121 (2024)
Tamburrini KC, Kodama S, Grisel S, Haon M, Nishiuchi T, Bissaro B, Kubo Y, Longhi S, Berrin JG
|
| RgGuinier |
6.4 |
nm |
| Dmax |
27.9 |
nm |
| VolumePorod |
199 |
nm3 |
|
|
|
|
|
|
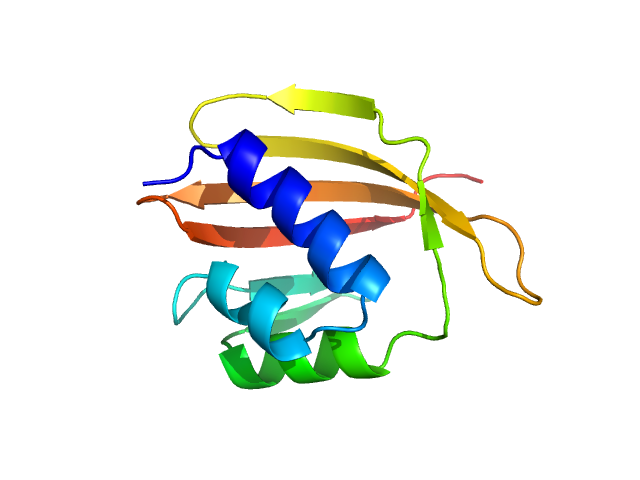
|
| Sample: |
SnoaL-like domain-containing protein monomer, 12 kDa Sphaerobacter thermophilus (strain … protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 3% glycerol, pH: 7.6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Nov 6
|
Elements of the C-terminal tail of a C-terminal domain homolog of the Orange Carotenoid Protein determining xanthophyll uptake from liposomes.
Biochim Biophys Acta Bioenerg 1865(3):149043 (2024)
Likkei K, Moldenhauer M, Tavraz NN, Egorkin NA, Slonimskiy YB, Maksimov EG, Sluchanko NN, Friedrich T
|
| RgGuinier |
1.5 |
nm |
| Dmax |
5.0 |
nm |
| VolumePorod |
18 |
nm3 |
|
|
|
|
|
|
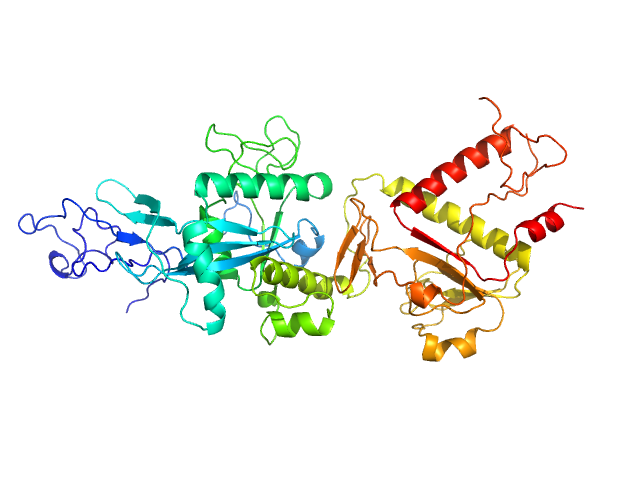
|
| Sample: |
Replicase polyprotein 1ab (non-structural protein 14) monomer, 60 kDa Severe acute respiratory … protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, 5 mM MgCl2, 2 mM β-mercaptoethanol, pH: 8.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2020 Dec 11
|
Despite the odds: formation of the SARS-CoV-2 methylation complex
Nucleic Acids Research (2024)
Matsuda A, Plewka J, Rawski M, Mourão A, Zajko W, Siebenmorgen T, Kresik L, Lis K, Jones A, Pachota M, Karim A, Hartman K, Nirwal S, Sonani R, Chykunova Y, Minia I, Mak P, Landthaler M, Nowotny M, Dubin G, Sattler M, Suder P, Popowicz G, Pyrć K, Czarna A
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.6 |
nm |
| VolumePorod |
80 |
nm3 |
|
|
|
|
|
|
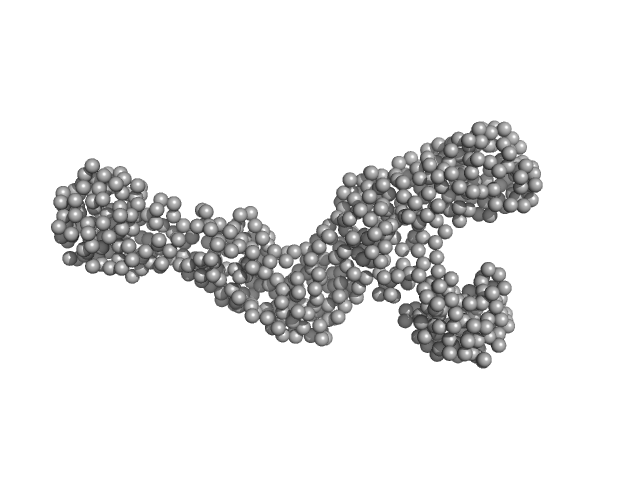
|
| Sample: |
Replicase polyprotein 1ab (non-structural protein 14) monomer, 60 kDa Severe acute respiratory … protein
Replicase polyprotein 1a (non-structural protein 10) monomer, 15 kDa Severe acute respiratory … protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, 5 mM MgCl2, 2 mM β-mercaptoethanol, pH: 8.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2020 Dec 11
|
Despite the odds: formation of the SARS-CoV-2 methylation complex
Nucleic Acids Research (2024)
Matsuda A, Plewka J, Rawski M, Mourão A, Zajko W, Siebenmorgen T, Kresik L, Lis K, Jones A, Pachota M, Karim A, Hartman K, Nirwal S, Sonani R, Chykunova Y, Minia I, Mak P, Landthaler M, Nowotny M, Dubin G, Sattler M, Suder P, Popowicz G, Pyrć K, Czarna A
|
| RgGuinier |
2.9 |
nm |
| Dmax |
12.2 |
nm |
| VolumePorod |
73 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Replicase polyprotein 1a (non-structural protein 10) monomer, 15 kDa Severe acute respiratory … protein
Replicase polyprotein 1ab (non-structural protein 16) monomer, 33 kDa Severe acute respiratory … protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, 5 mM MgCl2, 2 mM β-mercaptoethanol, pH: 8.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2020 Dec 11
|
Despite the odds: formation of the SARS-CoV-2 methylation complex
Nucleic Acids Research (2024)
Matsuda A, Plewka J, Rawski M, Mourão A, Zajko W, Siebenmorgen T, Kresik L, Lis K, Jones A, Pachota M, Karim A, Hartman K, Nirwal S, Sonani R, Chykunova Y, Minia I, Mak P, Landthaler M, Nowotny M, Dubin G, Sattler M, Suder P, Popowicz G, Pyrć K, Czarna A
|
| RgGuinier |
2.0 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
28 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Replicase polyprotein 1ab (non-structural protein 14) monomer, 60 kDa Severe acute respiratory … protein
Replicase polyprotein 1a (non-structural protein 10) monomer, 15 kDa Severe acute respiratory … protein
Replicase polyprotein 1ab (non-structural protein 16) monomer, 33 kDa Severe acute respiratory … protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, 5 mM MgCl2, 2 mM β-mercaptoethanol, pH: 8.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2020 Dec 11
|
Despite the odds: formation of the SARS-CoV-2 methylation complex
Nucleic Acids Research (2024)
Matsuda A, Plewka J, Rawski M, Mourão A, Zajko W, Siebenmorgen T, Kresik L, Lis K, Jones A, Pachota M, Karim A, Hartman K, Nirwal S, Sonani R, Chykunova Y, Minia I, Mak P, Landthaler M, Nowotny M, Dubin G, Sattler M, Suder P, Popowicz G, Pyrć K, Czarna A
|
| RgGuinier |
4.7 |
nm |
| Dmax |
11.4 |
nm |
| VolumePorod |
113 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Annexin A11 monomer, 54 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM KCl, 5 mM EDTA, 1 mM DTT, pH: 8.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 Jul 21
|
The structural properties of full-length annexin A11
Frontiers in Molecular Biosciences 11 (2024)
Dudas E, Tully M, Foldes T, Kelly G, Tartaglia G, Pastore A
|
| RgGuinier |
3.8 |
nm |
| Dmax |
15.9 |
nm |
| VolumePorod |
84 |
nm3 |
|
|