|
|
|
|
|
| Sample: |
Beta-amylase 2, chloroplastic tetramer, 229 kDa Arabidopsis thaliana protein
|
| Buffer: |
50 mM HEPES, 100 mM KCl, pH: 7 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Sep 17
|
Potassium cations expand the conformation ensemble of Arabidopsis thaliana β-amylase2 (BAM2).
MicroPubl Biol 2024 (2024)
Sholes A, Asongakap R, Jaconski S, Monroe J, Berndsen CE
|
| RgGuinier |
4.9 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
234 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Beta-amylase 2, chloroplastic tetramer, 229 kDa Arabidopsis thaliana protein
|
| Buffer: |
50 mM HEPES, 100 mM KCl, pH: 7 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Sep 17
|
Potassium cations expand the conformation ensemble of Arabidopsis thaliana β-amylase2 (BAM2).
MicroPubl Biol 2024 (2024)
Sholes A, Asongakap R, Jaconski S, Monroe J, Berndsen CE
|
| RgGuinier |
4.7 |
nm |
| Dmax |
15.1 |
nm |
| VolumePorod |
218 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Beta-amylase 2, chloroplastic tetramer, 229 kDa Arabidopsis thaliana protein
|
| Buffer: |
50 mM HEPES, 100 mM KCl, pH: 7 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Sep 17
|
Potassium cations expand the conformation ensemble of Arabidopsis thaliana β-amylase2 (BAM2).
MicroPubl Biol 2024 (2024)
Sholes A, Asongakap R, Jaconski S, Monroe J, Berndsen CE
|
| RgGuinier |
4.7 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
220 |
nm3 |
|
|
|
|
|
|
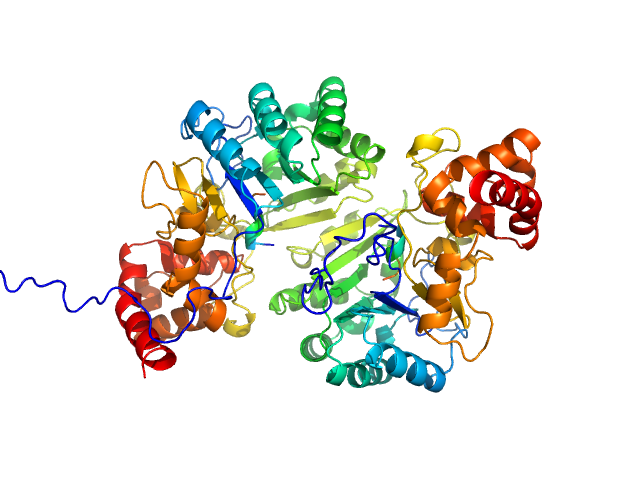
|
| Sample: |
Putative peptide biosynthesis protein YydG dimer, 80 kDa Bacillus subtilis (strain … protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Nov 30
|
Structural and mechanistic basis for RiPP epimerization by a radical SAM enzyme.
Nat Chem Biol (2023)
Kubiak X, Polsinelli I, Chavas LMG, Fyfe CD, Guillot A, Fradale L, Brewee C, Grimaldi S, Gerbaud G, Thureau A, Legrand P, Berteau O, Benjdia A
|
| RgGuinier |
2.9 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
113 |
nm3 |
|
|
|
|
|
|
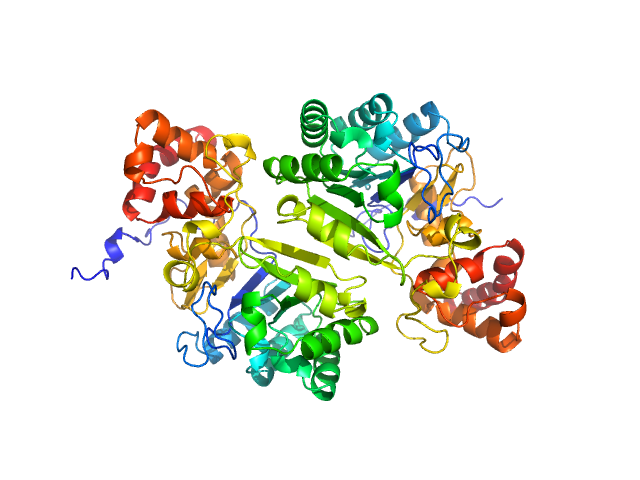
|
| Sample: |
Putative peptide biosynthesis protein YydG dimer, 80 kDa Bacillus subtilis (strain … protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Nov 29
|
Structural and mechanistic basis for RiPP epimerization by a radical SAM enzyme.
Nat Chem Biol (2023)
Kubiak X, Polsinelli I, Chavas LMG, Fyfe CD, Guillot A, Fradale L, Brewee C, Grimaldi S, Gerbaud G, Thureau A, Legrand P, Berteau O, Benjdia A
|
| RgGuinier |
2.9 |
nm |
| Dmax |
11.8 |
nm |
| VolumePorod |
115 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Beta-galactosidase-like enzyme dimer, 129 kDa Hamamotoa singularis protein
|
| Buffer: |
5 mM sodium phosphate, pH: 5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, Oak Ridge National Laboratory on 2018 Jun 25
|
Structural analysis and functional evaluation of the disordered ß–hexosyltransferase region from Hamamotoa (Sporobolomyces) singularis
Frontiers in Bioengineering and Biotechnology 11 (2023)
Dagher S, Vaishnav A, Stanley C, Meilleur F, Edwards B, Bruno-Bárcena J
|
| RgGuinier |
3.9 |
nm |
| Dmax |
12.4 |
nm |
| VolumePorod |
238 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Beta-galactosidase-like enzyme dimer, 129 kDa Hamamotoa singularis protein
|
| Buffer: |
5 mM sodium phosphate, pH: 5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, Oak Ridge National Laboratory on 2018 Jun 25
|
Structural analysis and functional evaluation of the disordered ß–hexosyltransferase region from Hamamotoa (Sporobolomyces) singularis
Frontiers in Bioengineering and Biotechnology 11 (2023)
Dagher S, Vaishnav A, Stanley C, Meilleur F, Edwards B, Bruno-Bárcena J
|
| RgGuinier |
3.7 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
236 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Tumor necrosis factor receptor superfamily member 17 monomer, 6 kDa Homo sapiens protein
Nanobody1 monomer, 14 kDa protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2022 Aug 28
|
Antigen-induced chimeric antigen receptor multimerization amplifies on-tumor cytotoxicity.
Signal Transduct Target Ther 8(1):445 (2023)
Sun Y, Yang XN, Yang SS, Lyu YZ, Zhang B, Liu KW, Li N, Cui JC, Huang GX, Liu CL, Xu J, Mi JQ, Chen Z, Fan XH, Chen SJ, Chen S
|
| RgGuinier |
3.0 |
nm |
| Dmax |
9.8 |
nm |
| VolumePorod |
52 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Tumor necrosis factor receptor superfamily member 17 monomer, 6 kDa Homo sapiens protein
Nanobody2 monomer, 14 kDa protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2022 Aug 28
|
Antigen-induced chimeric antigen receptor multimerization amplifies on-tumor cytotoxicity.
Signal Transduct Target Ther 8(1):445 (2023)
Sun Y, Yang XN, Yang SS, Lyu YZ, Zhang B, Liu KW, Li N, Cui JC, Huang GX, Liu CL, Xu J, Mi JQ, Chen Z, Fan XH, Chen SJ, Chen S
|
| RgGuinier |
2.0 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
24 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Tumor necrosis factor receptor superfamily member 17 monomer, 6 kDa Homo sapiens protein
Tandem nanobody monomer, 27 kDa protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2022 Aug 28
|
Antigen-induced chimeric antigen receptor multimerization amplifies on-tumor cytotoxicity.
Signal Transduct Target Ther 8(1):445 (2023)
Sun Y, Yang XN, Yang SS, Lyu YZ, Zhang B, Liu KW, Li N, Cui JC, Huang GX, Liu CL, Xu J, Mi JQ, Chen Z, Fan XH, Chen SJ, Chen S
|
| RgGuinier |
5.1 |
nm |
| Dmax |
23.3 |
nm |
| VolumePorod |
247 |
nm3 |
|
|