|
|
|
|
|
| Sample: |
Glutamate/aspartate import solute-binding protein monomer, 32 kDa Escherichia coli protein
|
| Buffer: |
100 mM NaCl, 20 mM NaPO4, 0.5 mM TCEP, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Sep 10
|
Structure-based screening of binding affinities via small-angle X-ray scattering
(2019)
Chen P, Masiewicz P, Perez K, Hennig J
|
| RgGuinier |
2.3 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
44 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Glutamate/aspartate import solute-binding protein monomer, 32 kDa Escherichia coli protein
|
| Buffer: |
100 mM NaCl, 20 mM NaPO4, 0.5 mM TCEP, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Sep 10
|
Structure-based screening of binding affinities via small-angle X-ray scattering
(2019)
Chen P, Masiewicz P, Perez K, Hennig J
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.4 |
nm |
| VolumePorod |
39 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Protein translocase subunit SecA dimer, 204 kDa Escherichia coli protein
|
| Buffer: |
20mM HEPES, 100mM NaCl, 1mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Jul 18
|
The C-terminal tail of the bacterial translocation ATPase SecA modulates its activity.
Elife 8 (2019)
Jamshad M, Knowles TJ, White SA, Ward DG, Mohammed F, Rahman KF, Wynne M, Hughes GW, Kramer G, Bukau B, Huber D
|
| RgGuinier |
4.2 |
nm |
| Dmax |
14.9 |
nm |
| VolumePorod |
424 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Protein translocase subunit SecA dimer, 199 kDa Escherichia coli protein
|
| Buffer: |
20mM HEPES, 100mM NaCl, 1mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Jul 18
|
The C-terminal tail of the bacterial translocation ATPase SecA modulates its activity.
Elife 8 (2019)
Jamshad M, Knowles TJ, White SA, Ward DG, Mohammed F, Rahman KF, Wynne M, Hughes GW, Kramer G, Bukau B, Huber D
|
| RgGuinier |
4.2 |
nm |
| Dmax |
14.8 |
nm |
| VolumePorod |
380 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Protein translocase subunit SecA dimer, 189 kDa Escherichia coli protein
|
| Buffer: |
20mM HEPES, 100mM NaCl, 1mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Jul 18
|
The C-terminal tail of the bacterial translocation ATPase SecA modulates its activity.
Elife 8 (2019)
Jamshad M, Knowles TJ, White SA, Ward DG, Mohammed F, Rahman KF, Wynne M, Hughes GW, Kramer G, Bukau B, Huber D
|
| RgGuinier |
4.5 |
nm |
| Dmax |
15.7 |
nm |
| VolumePorod |
398 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Fatty acid oxidation complex subunit alpha monomer, 81 kDa Escherichia coli protein
Fatty acid oxidation complex subunit alpha monomer, 81 kDa Escherichia coli protein
3-ketoacyl-CoA thiolase FadA (beta subunit) dimer, 82 kDa Escherichia coli protein
|
| Buffer: |
20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), 120 mM KCl, 2.5 mM DTT, pH: 7.2 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 May 30
|
Complementary substrate specificity and distinct quaternary assembly of the Escherichia coli aerobic and anaerobic beta-oxidation trifunctional enzyme complexes.
Biochem J (2019)
Sah-Teli SK, Hynönen MJ, Schmitz W, Geraets JA, Seitsonen J, Pedersen JS, Butcher SJ, Wierenga RK, Venkatesan R
|
| RgGuinier |
4.6 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
406 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Fatty acid oxidation complex subunit alpha monomer, 77 kDa Escherichia coli (strain … protein
Anaerobic Fatty acid oxidation complex subunit alpha monomer, 77 kDa Escherichia coli protein
Anaerobic Fatty acid oxidation complex subunit alpha monomer, 77 kDa Escherichia coli protein
Anaerobic Fatty acid oxidation complex subunit alpha monomer, 77 kDa Escherichia coli protein
Anaerobic 3-ketoacyl-CoA thiolase FadI beta subunit dimer, 96 kDa Escherichia coli protein
Anaerobic 3-ketoacyl-CoA thiolase FadI beta subunit dimer, 96 kDa Escherichia coli protein
|
| Buffer: |
50 mM Tris, 500 mM NaCl, 5% glycerol, 0.05% C12E9 (1-O-(n-Dodecyl)-nonaethyleneglycol), 2.5 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Sep 22
|
Complementary substrate specificity and distinct quaternary assembly of the Escherichia coli aerobic and anaerobic beta-oxidation trifunctional enzyme complexes.
Biochem J (2019)
Sah-Teli SK, Hynönen MJ, Schmitz W, Geraets JA, Seitsonen J, Pedersen JS, Butcher SJ, Wierenga RK, Venkatesan R
|
| RgGuinier |
6.2 |
nm |
| Dmax |
19.6 |
nm |
| VolumePorod |
856 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
DNA protection during starvation protein dodecamer, 224 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM Tris-HCl, 50 mM NaCl, 0.5 mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Nov 27
|
Protective Dps-DNA co-crystallization in stressed cells: an in vitro structural study by small-angle X-ray scattering and cryo-electron tomography.
FEBS Lett 593(12):1360-1371 (2019)
Dadinova LA, Chesnokov YM, Kamyshinsky RA, Orlov IA, Petoukhov MV, Mozhaev AA, Soshinskaya EY, Lazarev VN, Manuvera VA, Orekhov AS, Vasiliev AL, Shtykova EV
|
|
|
|
|
|
|
|
| Sample: |
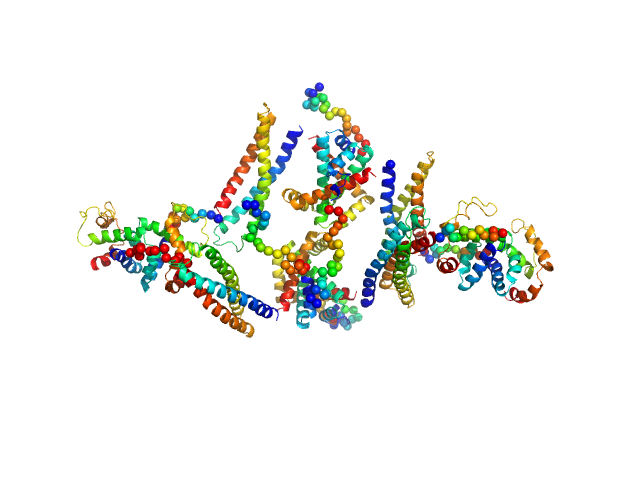
HP2042 form from Helicobacter pylori, N-terminal domain of syntaxin-1A from Rattus norvegicus, Trp repressor from Escherichia coli tetramer, 150 kDa Helicobacter pylori, Rattus … protein
|
| Buffer: |
20 mM Tris-HCl 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at Rigaku Nano-Viewer, Nara Institute of Science and Technology on 2016 Dec 8
|
Construction of a Quadrangular Tetramer and a Cage-Like Hexamer from Three-Helix Bundle-Linked Fusion Proteins.
ACS Synth Biol (2019)
Miyamoto T, Hayashi Y, Yoshida K, Watanabe H, Uchihashi T, Yonezawa K, Shimizu N, Kamikubo H, Hirota S
|
| RgGuinier |
5.1 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
379 |
nm3 |
|
|
|
|
|
|
|
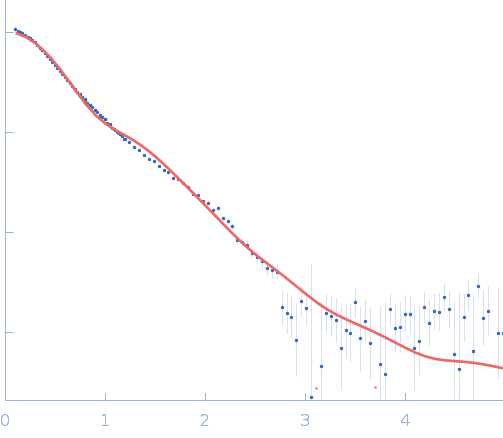
| Sample: |
Sulfite reductase [NADPH] flavoprotein alpha-component (Assimilatory NADPH-dependent sulfite reductase flavoprotein) monomer, 61 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM KPi, 100 mM NaCl, 1 mM EDTA, pH: 7.8 |
| Experiment: |
SANS
data collected at EQ-SANS (BL-6), Spallation Neutron Source on 2018 Jul 11
|
NADPH-dependent sulfite reductase flavoprotein adopts an extended conformation unique to this diflavin reductase
Journal of Structural Biology 205(2):170-179 (2019)
Tavolieri A, Murray D, Askenasy I, Pennington J, McGarry L, Stanley C, Stroupe M
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.6 |
nm |
| VolumePorod |
60 |
nm3 |
|
|