|
|
|
|
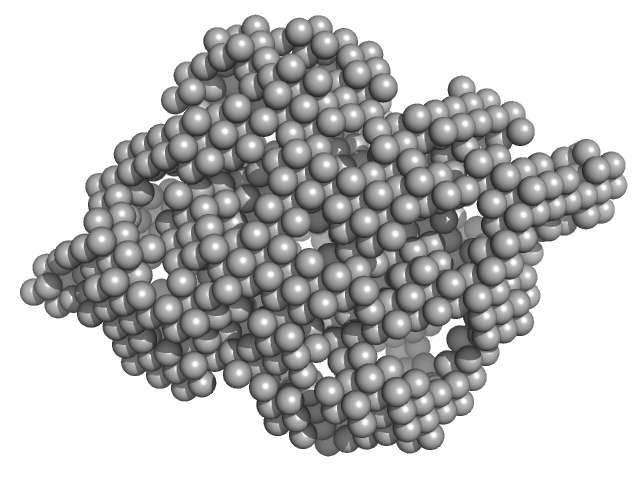
|
| Sample: |
Cysteine synthase A dimer, 71 kDa Escherichia coli protein
|
| Buffer: |
20 mM sodium phosphate, 85 mM NaCl, 2 mM EDTA, 10 mM 2-MCE, pH: 7.5 |
| Experiment: |
SAXS
data collected at Austrian SAXS beamline 5.2L, ELETTRA on 2016 Jun 1
|
Combination of SAXS and Protein Painting Discloses the Three-Dimensional Organization of the Bacterial Cysteine Synthase Complex, a Potential Target for Enhancers of Antibiotic Action.
Int J Mol Sci 20(20) (2019)
Rosa B, Marchetti M, Paredi G, Amenitsch H, Franko N, Benoni R, Giabbai B, De Marino MG, Mozzarelli A, Ronda L, Storici P, Campanini B, Bettati S
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
108 |
nm3 |
|
|
|
|
|
|
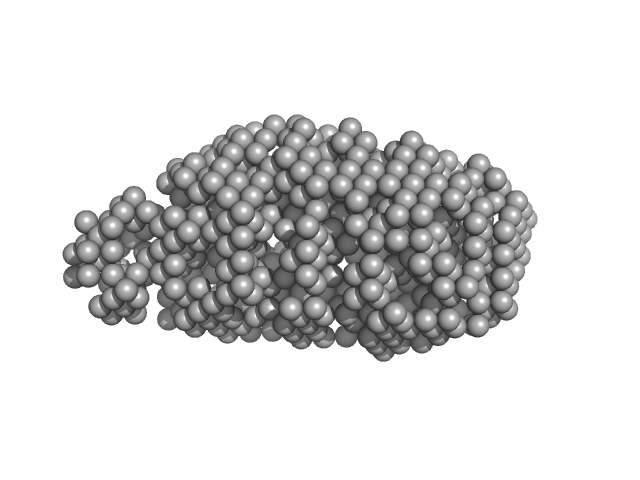
|
| Sample: |
Serine acetyltransferase hexamer, 177 kDa Escherichia coli protein
|
| Buffer: |
20 mM sodium phosphate, 85 mM NaCl, 2 mM EDTA, 10 mM 2-MCE, pH: 7.5 |
| Experiment: |
SAXS
data collected at Austrian SAXS beamline 5.2L, ELETTRA on 2016 Jun 1
|
Combination of SAXS and Protein Painting Discloses the Three-Dimensional Organization of the Bacterial Cysteine Synthase Complex, a Potential Target for Enhancers of Antibiotic Action.
Int J Mol Sci 20(20) (2019)
Rosa B, Marchetti M, Paredi G, Amenitsch H, Franko N, Benoni R, Giabbai B, De Marino MG, Mozzarelli A, Ronda L, Storici P, Campanini B, Bettati S
|
| RgGuinier |
3.9 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
280 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Cysteine synthase A (4-mer) tetramer, 143 kDa Escherichia coli protein
Serine acetyltransferase (6-mer) hexamer, 177 kDa Escherichia coli protein
|
| Buffer: |
20 mM sodium phosphate, 85 mM NaCl, 2 mM EDTA, 10 mM 2-MCE, pH: 7.5 |
| Experiment: |
SAXS
data collected at Austrian SAXS beamline 5.2L, ELETTRA on 2016 Jun 1
|
Combination of SAXS and Protein Painting Discloses the Three-Dimensional Organization of the Bacterial Cysteine Synthase Complex, a Potential Target for Enhancers of Antibiotic Action.
Int J Mol Sci 20(20) (2019)
Rosa B, Marchetti M, Paredi G, Amenitsch H, Franko N, Benoni R, Giabbai B, De Marino MG, Mozzarelli A, Ronda L, Storici P, Campanini B, Bettati S
|
| RgGuinier |
6.1 |
nm |
| Dmax |
22.0 |
nm |
| VolumePorod |
457 |
nm3 |
|
|
|
|
|
|
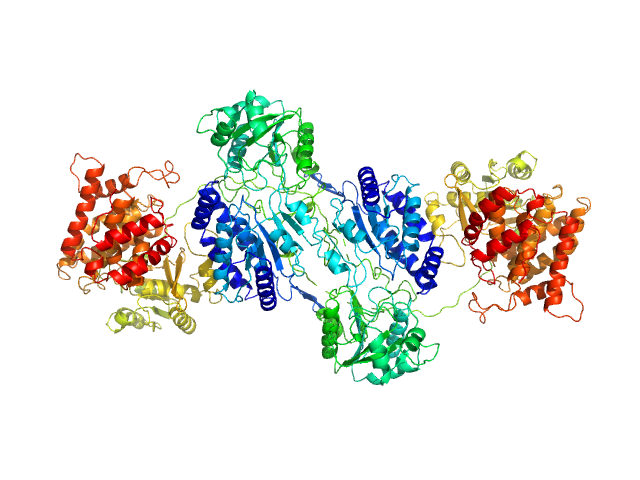
|
| Sample: |
F670E Aldehyde-alcohol dehydrogenase dimer, 192 kDa Escherichia coli protein
|
| Buffer: |
50 mM HEPES pH 7, 500 mM NaCl, 5% (v/v) glycerol, pH: 7 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Oct 4
|
Aldehyde-alcohol dehydrogenase forms a high-order spirosome architecture critical for its activity.
Nat Commun 10(1):4527 (2019)
Kim G, Azmi L, Jang S, Jung T, Hebert H, Roe AJ, Byron O, Song JJ
|
| RgGuinier |
5.0 |
nm |
| Dmax |
17.4 |
nm |
| VolumePorod |
260 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Zinc protease PqqL monomer, 102 kDa Escherichia coli protein
|
| Buffer: |
20 mM Tris HCl, 150 nM NaCl, 0.02 % NaN3, 5% glycerol, pH: 7.8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Jun 14
|
Protease-associated import systems are widespread in Gram-negative bacteria.
PLoS Genet 15(10):e1008435 (2019)
Grinter R, Leung PM, Wijeyewickrema LC, Littler D, Beckham S, Pike RN, Walker D, Greening C, Lithgow T
|
| RgGuinier |
4.0 |
nm |
| Dmax |
13.7 |
nm |
| VolumePorod |
207 |
nm3 |
|
|
|
|
|
|
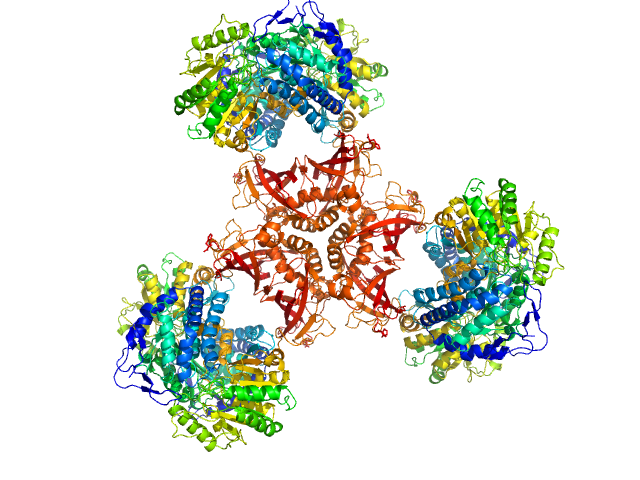
|
| Sample: |
Bifunctional protein PaaZ hexamer, 438 kDa Escherichia coli protein
|
| Buffer: |
25 mM HEPES, 50 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2015 Feb 24
|
Molecular basis for metabolite channeling in a ring opening enzyme of the phenylacetate degradation pathway.
Nat Commun 10(1):4127 (2019)
Sathyanarayanan N, Cannone G, Gakhar L, Katagihallimath N, Sowdhamini R, Ramaswamy S, Vinothkumar KR
|
| RgGuinier |
6.2 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
636 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Histidine-binding periplasmic protein monomer, 26 kDa Escherichia coli protein
|
| Buffer: |
100 mM NaCl, 20 mM NaPO4, 0.5 mM TCEP, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Sep 10
|
Structure-based screening of binding affinities via small-angle X-ray scattering
(2019)
Chen P, Masiewicz P, Perez K, Hennig J
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.0 |
nm |
| VolumePorod |
32 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Histidine-binding periplasmic protein monomer, 26 kDa Escherichia coli protein
|
| Buffer: |
100 mM NaCl, 20 mM NaPO4, 0.5 mM TCEP, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Sep 10
|
Structure-based screening of binding affinities via small-angle X-ray scattering
(2019)
Chen P, Masiewicz P, Perez K, Hennig J
|
| RgGuinier |
1.8 |
nm |
| Dmax |
5.7 |
nm |
| VolumePorod |
33 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Glutamine-binding periplasmic protein with hexahistidine tag monomer, 26 kDa Escherichia coli protein
|
| Buffer: |
100 mM NaCl, 20 mM NaPO4, 0.5 mM TCEP, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Sep 10
|
Structure-based screening of binding affinities via small-angle X-ray scattering
(2019)
Chen P, Masiewicz P, Perez K, Hennig J
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.2 |
nm |
| VolumePorod |
35 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Glutamine-binding periplasmic protein with hexahistidine tag monomer, 26 kDa Escherichia coli protein
|
| Buffer: |
100 mM NaCl, 20 mM NaPO4, 0.5 mM TCEP, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Sep 10
|
Structure-based screening of binding affinities via small-angle X-ray scattering
(2019)
Chen P, Masiewicz P, Perez K, Hennig J
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.1 |
nm |
| VolumePorod |
34 |
nm3 |
|
|