|
|
|
|
|
| Sample: |
Hemoglobin subunit beta monomer, 16 kDa Homo sapiens protein
Hemoglobin subunit alpha monomer, 15 kDa Homo sapiens protein
Protoporphyrin IX containing fe monomer, 1 kDa
|
| Buffer: |
100mM Sodium Phosphate buffer with 5% (w/v) PEG2000, pH: 7 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpoint 2.0, Institute of Biotechnology, Czech Academy of Sciences/Centre of Molecular Structure on 2022 Oct 6
|
Hemoglobin–PEG Interactions Probed by Small-Angle X-ray Scattering: Insights for Crystallization and Diagnostics Applications
The Journal of Physical Chemistry B (2024)
Baranova I, Angelova A, Stransky J, Andreasson J, Angelov B
|
| RgGuinier |
2.4 |
nm |
| Dmax |
6.5 |
nm |
| VolumePorod |
92 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Hemoglobin subunit alpha monomer, 15 kDa Homo sapiens protein
Hemoglobin subunit beta monomer, 16 kDa Homo sapiens protein
Protoporphyrin IX containing fe monomer, 1 kDa
|
| Buffer: |
100mM Sodium Phosphate buffer with 5% (w/v) PEG4000, pH: 7 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpoint 2.0, Institute of Biotechnology, Czech Academy of Sciences/Centre of Molecular Structure on 2022 Oct 6
|
Hemoglobin–PEG Interactions Probed by Small-Angle X-ray Scattering: Insights for Crystallization and Diagnostics Applications
The Journal of Physical Chemistry B (2024)
Baranova I, Angelova A, Stransky J, Andreasson J, Angelov B
|
| RgGuinier |
2.4 |
nm |
| Dmax |
6.1 |
nm |
| VolumePorod |
91 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Hemoglobin subunit alpha monomer, 15 kDa Homo sapiens protein
Hemoglobin subunit beta monomer, 16 kDa Homo sapiens protein
Protoporphyrin IX containing fe monomer, 1 kDa
|
| Buffer: |
100mM Sodium Phosphate buffer with 10% (w/v) PEG2000, pH: 7 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpoint 2.0, Institute of Biotechnology, Czech Academy of Sciences/Centre of Molecular Structure on 2022 Oct 6
|
Hemoglobin–PEG Interactions Probed by Small-Angle X-ray Scattering: Insights for Crystallization and Diagnostics Applications
The Journal of Physical Chemistry B (2024)
Baranova I, Angelova A, Stransky J, Andreasson J, Angelov B
|
| RgGuinier |
2.8 |
nm |
| Dmax |
11.3 |
nm |
| VolumePorod |
131 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Glutamate receptor ionotropic, NMDA 1 dimer, 193 kDa Homo sapiens protein
Glutamate receptor ionotropic, NMDA 2A dimer, 191 kDa Homo sapiens protein
|
| Buffer: |
150 mM NaCl, 0.1% digitonin, 5 µM Cholesteryl Hemisuccinate TRIS Salt, 0.1 mM CHAPSO, 50 µM EDTA,1 mM Gly/Glu, 20 mM HEPES, pH: 8 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2022 Dec 8
|
Structural basis for antibody-mediated NMDA receptor clustering and endocytosis in autoimmune encephalitis.
Nat Struct Mol Biol (2024)
Wang H, Xie C, Deng B, Ding J, Li N, Kou Z, Jin M, He J, Wang Q, Wen H, Zhang J, Zhou Q, Chen S, Chen X, Yuan TF, Zhu S
|
| RgGuinier |
6.6 |
nm |
| Dmax |
20.4 |
nm |
| VolumePorod |
1180 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Glutamate receptor ionotropic, NMDA 1 dimer, 193 kDa Homo sapiens protein
Glutamate receptor ionotropic, NMDA 2A dimer, 191 kDa Homo sapiens protein
Human derived autoantibody mAb2G7 heavy chain, mAb2G7 VH dimer, 103 kDa protein
Human derived autoantibody mAb2G7 light chain, mAb2G7 VL dimer, 51 kDa protein
|
| Buffer: |
150 mM NaCl, 0.1% digitonin, 5 µM Cholesteryl Hemisuccinate TRIS Salt, 0.1 mM CHAPSO, 50 µM EDTA,1 mM Gly/Glu, 20 mM HEPES, pH: 8 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2022 Dec 8
|
Structural basis for antibody-mediated NMDA receptor clustering and endocytosis in autoimmune encephalitis.
Nat Struct Mol Biol (2024)
Wang H, Xie C, Deng B, Ding J, Li N, Kou Z, Jin M, He J, Wang Q, Wen H, Zhang J, Zhou Q, Chen S, Chen X, Yuan TF, Zhu S
|
| RgGuinier |
7.7 |
nm |
| Dmax |
25.4 |
nm |
| VolumePorod |
1260 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Glutamate receptor ionotropic, NMDA 1 dimer, 193 kDa Homo sapiens protein
Glutamate receptor ionotropic, NMDA 2A dimer, 191 kDa Homo sapiens protein
Human derived autoantibody mAb5F6 heavy chain, mAb5F6 VH dimer, 104 kDa Homo sapiens protein
Human derived autoantibody mAb5F6 light chain, mAb5F6 VL dimer, 52 kDa protein
|
| Buffer: |
150 mM NaCl, 0.1% digitonin, 5 µM Cholesteryl Hemisuccinate TRIS Salt, 0.1 mM CHAPSO, 50 µM EDTA,1 mM Gly/Glu, 20 mM HEPES, pH: 8 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2022 Dec 8
|
Structural basis for antibody-mediated NMDA receptor clustering and endocytosis in autoimmune encephalitis.
Nat Struct Mol Biol (2024)
Wang H, Xie C, Deng B, Ding J, Li N, Kou Z, Jin M, He J, Wang Q, Wen H, Zhang J, Zhou Q, Chen S, Chen X, Yuan TF, Zhu S
|
| RgGuinier |
9.9 |
nm |
| Dmax |
31.7 |
nm |
| VolumePorod |
2500 |
nm3 |
|
|
|
|
|
|
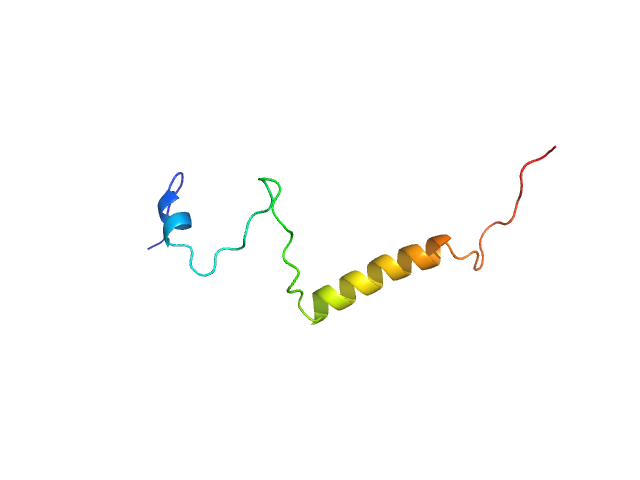
|
| Sample: |
Isoform Short of Small EDRK-rich factor 1 monomer, 7 kDa Homo sapiens protein
|
| Buffer: |
Sodium phosphate buffer, pH: 7.4 |
| Experiment: |
SAXS
data collected at TPS13A, NSRRC on 2021 Mar 11
|
Binding structures of SERF1a with NT17-polyQ peptides of huntingtin exon 1 revealed by SEC-SWAXS, NMR and molecular simulation.
IUCrJ (2024)
Lin TC, Shih O, Tsai TY, Yeh YQ, Liao KF, Mansel BW, Shiu YJ, Chang CF, Su AC, Chen YR, Jeng US
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
9 |
nm3 |
|
|
|
|
|
|
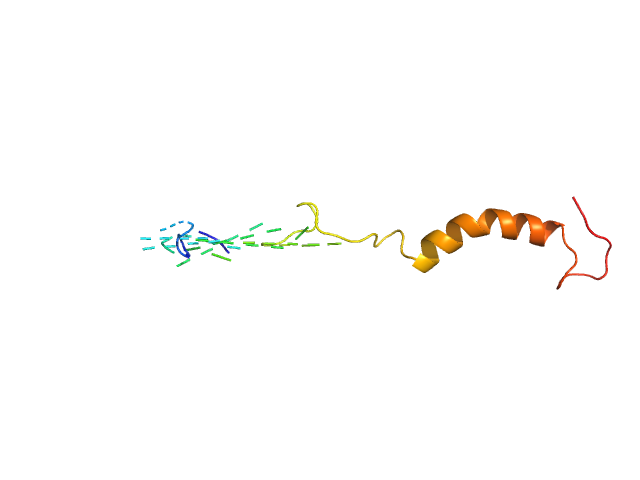
|
| Sample: |
Isoform Short of Small EDRK-rich factor 1 monomer, 7 kDa Homo sapiens protein
NT17 dimer, 4 kDa synthetic construct protein
|
| Buffer: |
Sodium phosphate buffer, pH: 7.4 |
| Experiment: |
SAXS
data collected at TPS13A, NSRRC on 2021 Oct 21
|
Binding structures of SERF1a with NT17-polyQ peptides of huntingtin exon 1 revealed by SEC-SWAXS, NMR and molecular simulation.
IUCrJ (2024)
Lin TC, Shih O, Tsai TY, Yeh YQ, Liao KF, Mansel BW, Shiu YJ, Chang CF, Su AC, Chen YR, Jeng US
|
|
|
|
|
|
|
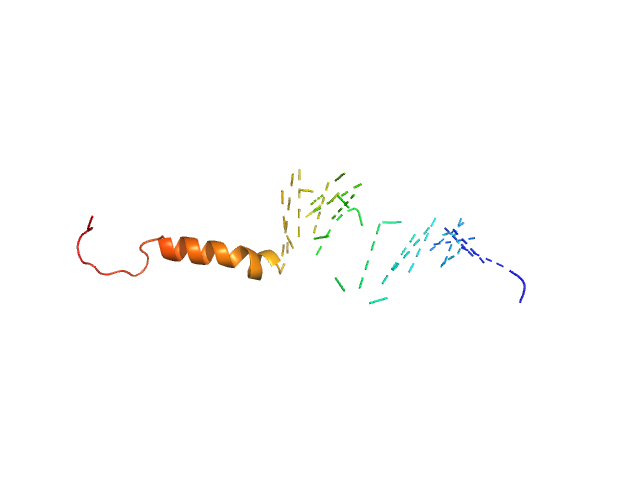
|
| Sample: |
Isoform Short of Small EDRK-rich factor 1 monomer, 7 kDa Homo sapiens protein
HTT3 monomer, 4 kDa synthetic construct protein
|
| Buffer: |
Sodium phosphate buffer, pH: 7.4 |
| Experiment: |
SAXS
data collected at TPS13A, NSRRC on 2021 May 20
|
Binding structures of SERF1a with NT17-polyQ peptides of huntingtin exon 1 revealed by SEC-SWAXS, NMR and molecular simulation.
IUCrJ (2024)
Lin TC, Shih O, Tsai TY, Yeh YQ, Liao KF, Mansel BW, Shiu YJ, Chang CF, Su AC, Chen YR, Jeng US
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.2 |
nm |
| VolumePorod |
9 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
DNA repair protein RAD52 homolog decamer, 234 kDa Homo sapiens protein
|
| Buffer: |
20 mM Bis-Tris, 10% glycerol, 400 mM NaCl, 100 mM KCl, 1 mM EDTA, pH: 6 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS 1000, Eppley Structural Biology Facility, University of Nebraska Medical Center on 2014 Jul 25
|
A glimpse into the hidden world of the flexible C-terminal protein binding domains of human RAD52
Journal of Structural Biology 216(3):108115 (2024)
Struble L, Lovelace J, Borgstahl G
|
| RgGuinier |
4.1 |
nm |
| Dmax |
11.8 |
nm |
| VolumePorod |
375 |
nm3 |
|
|