|
|
|
|

|
| Sample: |
GAGE6 60bp dsDNA oligo monomer, 37 kDa Homo sapiens DNA
|
| Buffer: |
150 mM KCl, 20 mM HEPES, 5% glycerol, 5 mM MgCl2, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2023 Mar 14
|
Probing the statistics of sequence-dependent DNA conformations in solution using SAXS
Heidar Koning
|
| RgGuinier |
4.7 |
nm |
| Dmax |
20.7 |
nm |
| VolumePorod |
101 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
GAGE6_1 dimer, 37 kDa Homo sapiens DNA
|
| Buffer: |
150 mM KCl, 20 mM HEPES, 5% glycerol, 5 mM MgCl2, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2023 Nov 28
|
Probing the statistics of sequence-dependent DNA conformations in solution using SAXS
Heidar Koning
|
| RgGuinier |
5.1 |
nm |
| Dmax |
19.2 |
nm |
| VolumePorod |
67 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
GAGE6_2 dimer, 37 kDa Homo sapiens DNA
|
| Buffer: |
150 mM KCl, 20 mM HEPES, 5% glycerol, 5 mM MgCl2, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2023 Nov 28
|
Probing the statistics of sequence-dependent DNA conformations in solution using SAXS
Heidar Koning
|
| RgGuinier |
5.2 |
nm |
| Dmax |
18.4 |
nm |
| VolumePorod |
72 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
GAGE6_3 dimer, 37 kDa Homo sapiens DNA
|
| Buffer: |
150 mM KCl, 20 mM HEPES, 5% glycerol, 5 mM MgCl2, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2023 Nov 28
|
Probing the statistics of sequence-dependent DNA conformations in solution using SAXS
Heidar Koning
|
| RgGuinier |
5.0 |
nm |
| Dmax |
21.4 |
nm |
| VolumePorod |
80 |
nm3 |
|
|
|
|
|
|
|
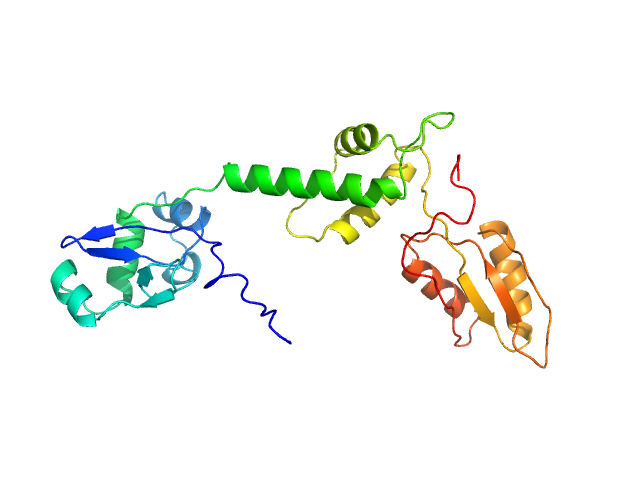
| Sample: |
Ribosome maturation protein SBDS monomer, 29 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris pH 8.0, 10% glycerol, 300 mM NaCl, 5 mM MgCl2., pH: |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Apr 6
|
Structural Implications of Missense Point Mutations in Shwachman–Bodian–Diamond Syndrome Protein (SBDS): A Combined SAXS/MD Investigation
ACS Omega (2025)
Mattiotti G, Nanna V, Giulini M, Alberga D, Mangiatordi G, Sánchez-Puig N, Saviano M, Tubiana L, Potestio R, Lattanzi G, Siliqi D
|
| RgGuinier |
3.0 |
nm |
| Dmax |
11.4 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
|
|
|
|
|
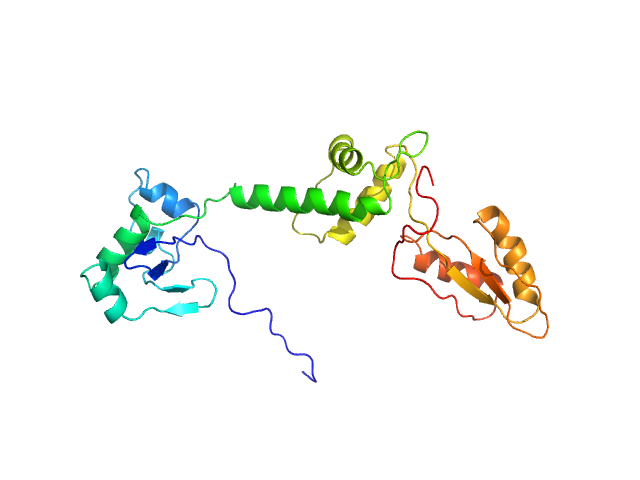
| Sample: |
Ribosome maturation protein SBDS (R19Q) monomer, 29 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris pH 8.0, 10% glycerol, 300 mM NaCl, 5 mM MgCl2., pH: |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Apr 6
|
Structural Implications of Missense Point Mutations in Shwachman–Bodian–Diamond Syndrome Protein (SBDS): A Combined SAXS/MD Investigation
ACS Omega (2025)
Mattiotti G, Nanna V, Giulini M, Alberga D, Mangiatordi G, Sánchez-Puig N, Saviano M, Tubiana L, Potestio R, Lattanzi G, Siliqi D
|
| RgGuinier |
2.9 |
nm |
| Dmax |
12.2 |
nm |
| VolumePorod |
46 |
nm3 |
|
|
|
|
|
|
|
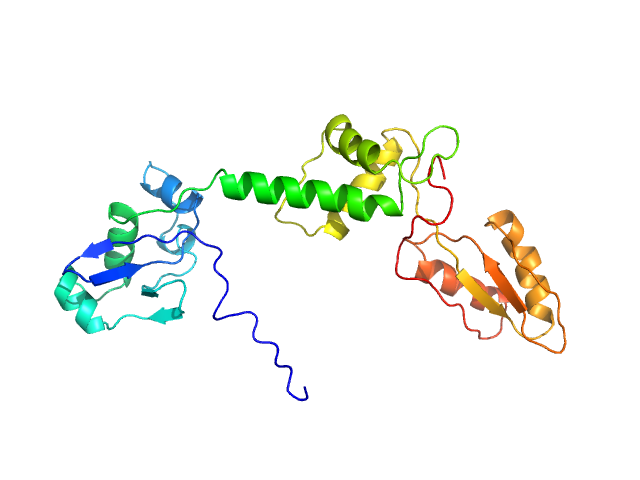
| Sample: |
Ribosome maturation protein SBDS (I167T) monomer, 29 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris pH 8.0, 10% glycerol, 300 mM NaCl, 5 mM MgCl2., pH: |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Apr 6
|
Structural Implications of Missense Point Mutations in Shwachman–Bodian–Diamond Syndrome Protein (SBDS): A Combined SAXS/MD Investigation
ACS Omega (2025)
Mattiotti G, Nanna V, Giulini M, Alberga D, Mangiatordi G, Sánchez-Puig N, Saviano M, Tubiana L, Potestio R, Lattanzi G, Siliqi D
|
| RgGuinier |
2.9 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
44 |
nm3 |
|
|
|
|
|
|
|
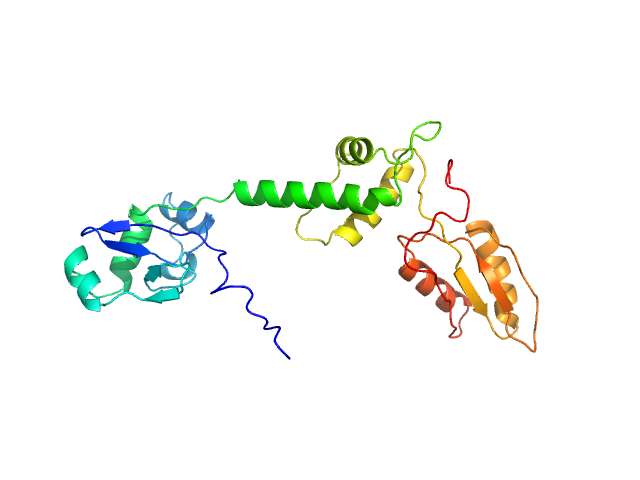
| Sample: |
Ribosome maturation protein SBDS (R175W) monomer, 29 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris pH 8.0, 10% glycerol, 300 mM NaCl, 5 mM MgCl2., pH: |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Apr 6
|
Structural Implications of Missense Point Mutations in Shwachman–Bodian–Diamond Syndrome Protein (SBDS): A Combined SAXS/MD Investigation
ACS Omega (2025)
Mattiotti G, Nanna V, Giulini M, Alberga D, Mangiatordi G, Sánchez-Puig N, Saviano M, Tubiana L, Potestio R, Lattanzi G, Siliqi D
|
| RgGuinier |
3.1 |
nm |
| Dmax |
11.6 |
nm |
| VolumePorod |
48 |
nm3 |
|
|
|
|
|
|
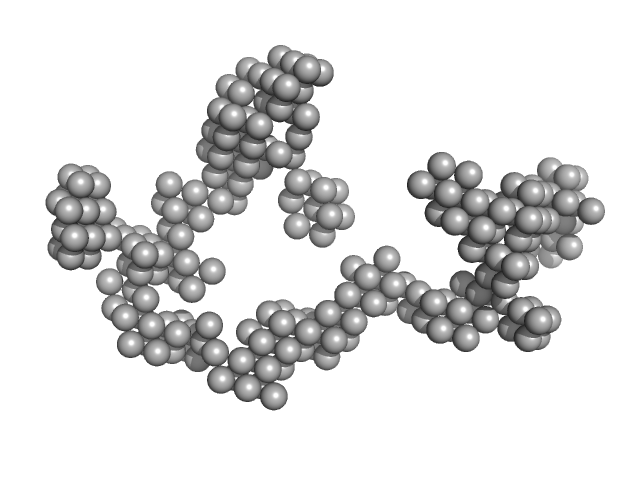
|
| Sample: |
RNA component of 7SK nuclear ribonucleoprotein monomer, 107 kDa Homo sapiens RNA
|
| Buffer: |
10 mM Bis-Tris, 100 mM NaCl, 15 mM KCl, 5% Glycerol, 3 mM MgCl2, pH: 5.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2024 Jun 21
|
Solution Structure Determination and Biophysical Studies of 7SK RNP and 7SL SRP RNAs.
RNA (2025)
D'Souza MH, Pereira HS, Tersteeg S, Vasudeva G, Siddiqui MQ, Agunlejika T, Kerr L, Girodat D, Patel T
|
| RgGuinier |
8.0 |
nm |
| Dmax |
23.2 |
nm |
| VolumePorod |
600 |
nm3 |
|
|
|
|
|
|
|
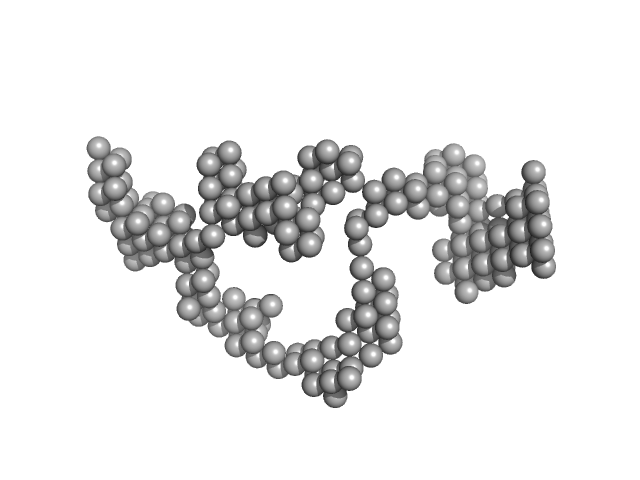
| Sample: |
RNA component of 7SK nuclear ribonucleoprotein monomer, 107 kDa Homo sapiens RNA
|
| Buffer: |
10 mM Bis-Tris, 100 mM NaCl, 15 mM KCl, 5% Glycerol, 3 mM MgCl2, pH: 5.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2024 Jun 21
|
Solution Structure Determination and Biophysical Studies of 7SK RNP and 7SL SRP RNAs.
RNA (2025)
D'Souza MH, Pereira HS, Tersteeg S, Vasudeva G, Siddiqui MQ, Agunlejika T, Kerr L, Girodat D, Patel T
|
| RgGuinier |
7.9 |
nm |
| Dmax |
24.1 |
nm |
| VolumePorod |
290 |
nm3 |
|
|