|
|
|
|
|
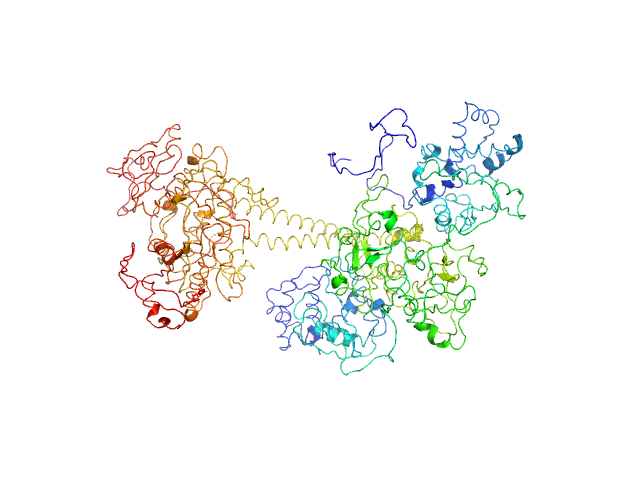
| Sample: |
Cyclic di-AMP binding protein (Putative regulatory, ligand-binding protein) dimer, 38 kDa Streptomyces venezuelae protein
|
| Buffer: |
200 mM NaCl, 30 mM NaPi, 5% (v/v) glycerol, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Nov 4
|
c-di-AMP hydrolysis by a novel type of phosphodiesterase promotes differentiation of multicellular bacteria
(2019)
Latoscha A, Drexler D, Al-Bassam M, Kaever V, Findlay K, Witte G, Tschowri N
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.2 |
nm |
| VolumePorod |
69 |
nm3 |
|
|
|
|
|
|
|
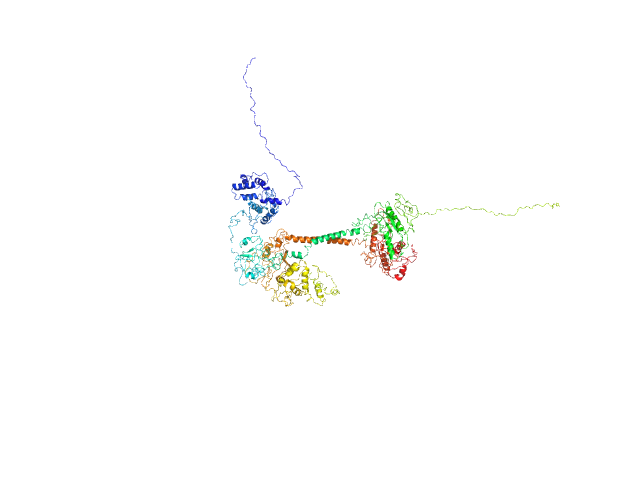
| Sample: |
Soluble guanylyl cyclase alpha-1 subunit monomer, 78 kDa Manduca sexta protein
Soluble guanylyl cyclase beta-1 subunit monomer, 68 kDa Manduca sexta protein
|
| Buffer: |
50 mM KH2PO4, 150 mM NaCl, 2% glycerol,, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Dec 3
|
Allosteric activation of the nitric oxide receptor soluble guanylate cyclase mapped by cryo-electron microscopy.
Elife 8 (2019)
Horst BG, Yokom AL, Rosenberg DJ, Morris KL, Hammel M, Hurley JH, Marletta MA
|
| RgGuinier |
4.3 |
nm |
| Dmax |
13.3 |
nm |
| VolumePorod |
230 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Soluble guanylyl cyclase alpha-1 subunit monomer, 78 kDa Manduca sexta protein
Soluble guanylyl cyclase beta-1 subunit monomer, 68 kDa Manduca sexta protein
|
| Buffer: |
50 mM KH2PO4,150 mM NaCl, 2% glycerol, 500 µM NO,, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Dec 3
|
Allosteric activation of the nitric oxide receptor soluble guanylate cyclase mapped by cryo-electron microscopy.
Elife 8 (2019)
Horst BG, Yokom AL, Rosenberg DJ, Morris KL, Hammel M, Hurley JH, Marletta MA
|
| RgGuinier |
4.4 |
nm |
| Dmax |
14.2 |
nm |
| VolumePorod |
218 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
DNA-directed RNA polymerase subunit delta monomer, 20 kDa Bacillus subtilis protein
|
| Buffer: |
20 mM Phosphate buffer, 10 mM NaCl, 0.05% NaN3, pH: 6.6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Oct 3
|
Quantitative Conformational Analysis of Functionally Important Electrostatic Interactions in the Intrinsically Disordered Region of Delta Subunit of Bacterial RNA Polymerase.
J Am Chem Soc (2019)
Kuban V, Srb P, Stegnerova H, Padrta P, Zachrdla M, Jasenakova Z, Šanderová H, Vítovská D, Krasny L, Koval T, Dohnalek J, Ziemska-Legi Cka J, Grynberg M, Jarnot P, Gruca A, Jensen MR, Blackledge M, Zidek L
|
| RgGuinier |
3.5 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
38 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
DNA-directed RNA polymerase subunit delta monomer, 20 kDa Bacillus subtilis protein
|
| Buffer: |
20 mM Phosphate buffer, 200 mM NaCl, 0.05% NaN3, pH: 6.6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Oct 3
|
Quantitative Conformational Analysis of Functionally Important Electrostatic Interactions in the Intrinsically Disordered Region of Delta Subunit of Bacterial RNA Polymerase.
J Am Chem Soc (2019)
Kuban V, Srb P, Stegnerova H, Padrta P, Zachrdla M, Jasenakova Z, Šanderová H, Vítovská D, Krasny L, Koval T, Dohnalek J, Ziemska-Legi Cka J, Grynberg M, Jarnot P, Gruca A, Jensen MR, Blackledge M, Zidek L
|
| RgGuinier |
3.9 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
56 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
DNA-directed RNA polymerase subunit delta monomer, 20 kDa Bacillus subtilis protein
|
| Buffer: |
20 mM Phosphate buffer, 400 mM NaCl, 0.05% NaN3, pH: 6.6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Oct 3
|
Quantitative Conformational Analysis of Functionally Important Electrostatic Interactions in the Intrinsically Disordered Region of Delta Subunit of Bacterial RNA Polymerase.
J Am Chem Soc (2019)
Kuban V, Srb P, Stegnerova H, Padrta P, Zachrdla M, Jasenakova Z, Šanderová H, Vítovská D, Krasny L, Koval T, Dohnalek J, Ziemska-Legi Cka J, Grynberg M, Jarnot P, Gruca A, Jensen MR, Blackledge M, Zidek L
|
| RgGuinier |
4.2 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
63 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
DNA-directed RNA polymerase subunit delta monomer, 20 kDa Bacillus subtilis protein
|
| Buffer: |
20 mM Phosphate buffer, 800 mM NaCl, 0.05% NaN3, pH: 6.6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Oct 3
|
Quantitative Conformational Analysis of Functionally Important Electrostatic Interactions in the Intrinsically Disordered Region of Delta Subunit of Bacterial RNA Polymerase.
J Am Chem Soc (2019)
Kuban V, Srb P, Stegnerova H, Padrta P, Zachrdla M, Jasenakova Z, Šanderová H, Vítovská D, Krasny L, Koval T, Dohnalek J, Ziemska-Legi Cka J, Grynberg M, Jarnot P, Gruca A, Jensen MR, Blackledge M, Zidek L
|
| RgGuinier |
4.5 |
nm |
| Dmax |
21.0 |
nm |
| VolumePorod |
77 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
DNA-directed RNA polymerase subunit delta - mutant monomer, 20 kDa Bacillus subtilis protein
|
| Buffer: |
20 mM Phosphate buffer, 10 mM NaCl, 0.05% NaN3, pH: 6.6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Oct 3
|
Quantitative Conformational Analysis of Functionally Important Electrostatic Interactions in the Intrinsically Disordered Region of Delta Subunit of Bacterial RNA Polymerase.
J Am Chem Soc (2019)
Kuban V, Srb P, Stegnerova H, Padrta P, Zachrdla M, Jasenakova Z, Šanderová H, Vítovská D, Krasny L, Koval T, Dohnalek J, Ziemska-Legi Cka J, Grynberg M, Jarnot P, Gruca A, Jensen MR, Blackledge M, Zidek L
|
| RgGuinier |
4.3 |
nm |
| Dmax |
19.5 |
nm |
| VolumePorod |
58 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
DNA-directed RNA polymerase subunit delta - mutant monomer, 20 kDa Bacillus subtilis protein
|
| Buffer: |
20 mM Phosphate buffer, 200 mM NaCl, 0.05% NaN3, pH: 6.6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Oct 3
|
Quantitative Conformational Analysis of Functionally Important Electrostatic Interactions in the Intrinsically Disordered Region of Delta Subunit of Bacterial RNA Polymerase.
J Am Chem Soc (2019)
Kuban V, Srb P, Stegnerova H, Padrta P, Zachrdla M, Jasenakova Z, Šanderová H, Vítovská D, Krasny L, Koval T, Dohnalek J, Ziemska-Legi Cka J, Grynberg M, Jarnot P, Gruca A, Jensen MR, Blackledge M, Zidek L
|
| RgGuinier |
4.6 |
nm |
| Dmax |
22.0 |
nm |
| VolumePorod |
76 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
DNA-directed RNA polymerase subunit delta - mutant monomer, 20 kDa Bacillus subtilis protein
|
| Buffer: |
20 mM Phosphate buffer, 400 mM NaCl, 0.05% NaN3, pH: 6.6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Oct 3
|
Quantitative Conformational Analysis of Functionally Important Electrostatic Interactions in the Intrinsically Disordered Region of Delta Subunit of Bacterial RNA Polymerase.
J Am Chem Soc (2019)
Kuban V, Srb P, Stegnerova H, Padrta P, Zachrdla M, Jasenakova Z, Šanderová H, Vítovská D, Krasny L, Koval T, Dohnalek J, Ziemska-Legi Cka J, Grynberg M, Jarnot P, Gruca A, Jensen MR, Blackledge M, Zidek L
|
| RgGuinier |
4.5 |
nm |
| Dmax |
24.0 |
nm |
| VolumePorod |
78 |
nm3 |
|
|