|
|
|
|
|
| Sample: |
Candidate inclusion membrane protein (MBP-tagged) tetramer, 235 kDa Chlamydia trachomatis serovar … protein
|
| Buffer: |
20 mM Tris-HCl, 50 mM NaCl, 1 mM EDTA, 2% glycerol, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2023 Feb 15
|
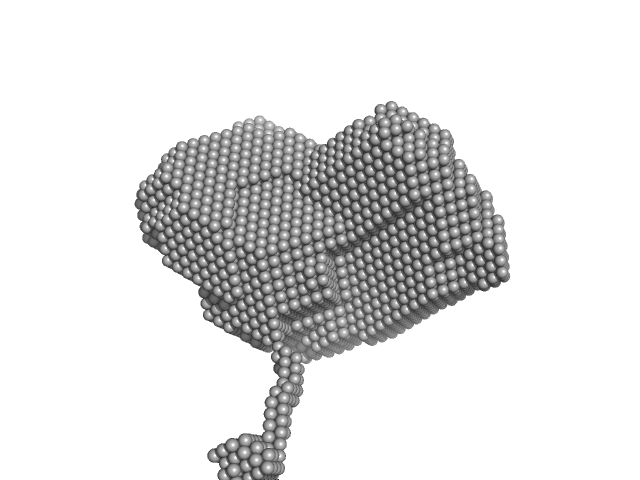
Tetramer formation of CpoS facilitates Inc-Inc interactions during Chlamydia trachomatis infection
Zhen Xu
|
| RgGuinier |
7.1 |
nm |
| Dmax |
30.3 |
nm |
| VolumePorod |
625 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Group IIC Intron Domain 1 monomer, 89 kDa Oceanobacillus iheyensis RNA
|
| Buffer: |
10 mM MgCl2, 5 mM Na-MES, pH: 6.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Jul 7
|
Dynamic assembly of a large multidomain ribozyme visualized by cryo-electron microscopy.
Nat Commun 16(1):10195 (2025)
Jadhav S, Maiorca M, Manigrasso J, Saha S, Rakitch A, Muscat S, Mulvaney T, De Vivo M, Topf M, Marcia M
|
| RgGuinier |
3.8 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
188 |
nm3 |
|
|
|
|
|
|
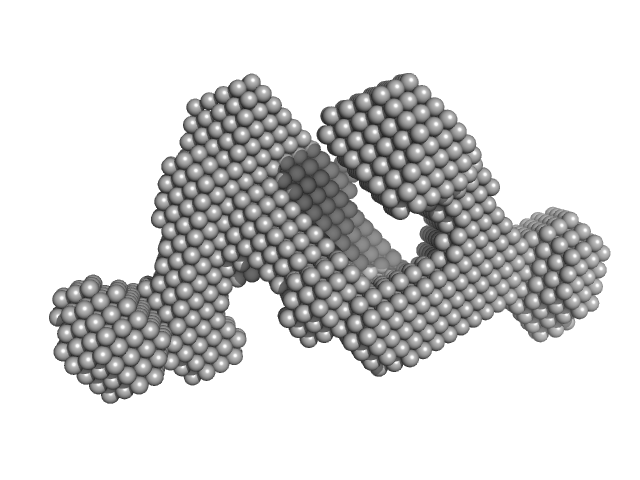
|
| Sample: |
Group IIC Intron Domain 1, 2 & 3 monomer, 103 kDa Oceanobacillus iheyensis RNA
|
| Buffer: |
10 mM MgCl2, 5 mM Na-MES, pH: 6.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Sep 2
|
Dynamic assembly of a large multidomain ribozyme visualized by cryo-electron microscopy.
Nat Commun 16(1):10195 (2025)
Jadhav S, Maiorca M, Manigrasso J, Saha S, Rakitch A, Muscat S, Mulvaney T, De Vivo M, Topf M, Marcia M
|
| RgGuinier |
3.6 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
195 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Group IIC Intron Domain 1 & 2 monomer, 96 kDa Oceanobacillus iheyensis RNA
|
| Buffer: |
10 mM MgCl2, 5 mM Na-MES, pH: 6.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Sep 2
|
Dynamic assembly of a large multidomain ribozyme visualized by cryo-electron microscopy.
Nat Commun 16(1):10195 (2025)
Jadhav S, Maiorca M, Manigrasso J, Saha S, Rakitch A, Muscat S, Mulvaney T, De Vivo M, Topf M, Marcia M
|
| RgGuinier |
3.6 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
179 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Group IIC intron Domain 1, 2, 3 & 4 monomer, 125 kDa Oceanobacillus iheyensis RNA
|
| Buffer: |
10 mM MgCl2, 5 mM Na-MES, pH: 6.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Sep 2
|
Dynamic assembly of a large multidomain ribozyme visualized by cryo-electron microscopy.
Nat Commun 16(1):10195 (2025)
Jadhav S, Maiorca M, Manigrasso J, Saha S, Rakitch A, Muscat S, Mulvaney T, De Vivo M, Topf M, Marcia M
|
| RgGuinier |
4.2 |
nm |
| Dmax |
12.6 |
nm |
| VolumePorod |
247 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Estrogen receptor alpha dimer, 57 kDa Melanotaenia fluviatilis protein
|
| Buffer: |
20 mM Tris-HCl pH 7.5, 150 mM NaCl, 5% glycerol, 2 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioSAXS, Australian Synchrotron on 2024 Nov 9
|
A ternary switch model governing ERα ligand binding domain conformation.
Nat Commun 16(1):10363 (2025)
McDougal DP, Pederick JL, Novick SJ, Jovcevski B, Warrender AK, Pascal BD, Griffin PR, Bruning JB
|
| RgGuinier |
2.5 |
nm |
| Dmax |
9.2 |
nm |
| VolumePorod |
72 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Ubiquitinating/deubiquitinating enzyme SdeA monomer, 39 kDa Legionella pneumophila subsp. … protein
|
| Buffer: |
25 mM Tris-HCl, 200 mM NaCl, 5 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Dec 4
|
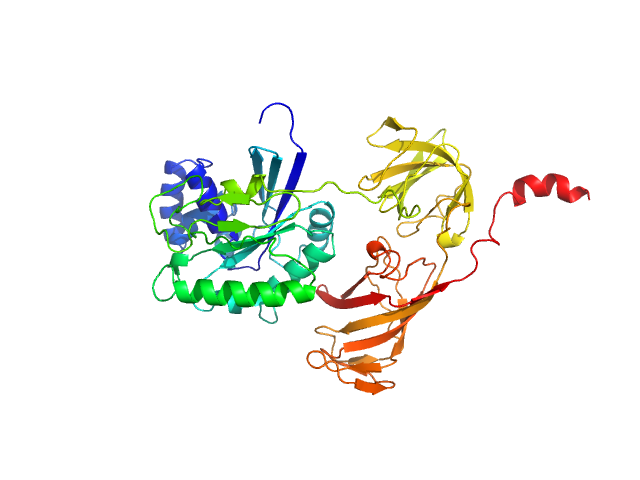
Structural Basis for Membrane Targeting and Secretion of Legionella SidE Ubiquitin Ligases
Ahmed Mohammed
|
| RgGuinier |
3.9 |
nm |
| Dmax |
15.7 |
nm |
| VolumePorod |
62 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Type 4 adapter protein IcmS monomer, 14 kDa Legionella pneumophila subsp. … protein
Type 4 adapter protein IcmW (S9A) monomer, 17 kDa Legionella pneumophila subsp. … protein
Type 4 coupling protein DotL monomer, 15 kDa Legionella pneumophila subsp. … protein
|
| Buffer: |
25 mM Tris-HCl, 200 mM NaCl, 5 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Dec 4
|
Structural Basis for Membrane Targeting and Secretion of Legionella SidE Ubiquitin Ligases
Ahmed Mohammed
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.6 |
nm |
| VolumePorod |
71 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Ubiquitinating/deubiquitinating enzyme SdeA monomer, 39 kDa Legionella pneumophila subsp. … protein
Type 4 adapter protein IcmS monomer, 14 kDa Legionella pneumophila subsp. … protein
Type 4 adapter protein IcmW (S9A) monomer, 17 kDa Legionella pneumophila subsp. … protein
Type 4 coupling protein DotL monomer, 15 kDa Legionella pneumophila subsp. … protein
|
| Buffer: |
25 mM Tris-HCl, 200 mM NaCl, 5 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Dec 4
|
Structural Basis for Membrane Targeting and Secretion of Legionella SidE Ubiquitin Ligases
Ahmed Mohammed
|
| RgGuinier |
3.8 |
nm |
| Dmax |
14.4 |
nm |
| VolumePorod |
166 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Mammalian translation elongation factor eEF1A2 monomer, 50 kDa protein
|
| Buffer: |
25 mM Tris HCl, 150 mM NaCl, 6 mM βME, 20% glycerol, 0.01mM GDP,, pH: 7.5 |
| Experiment: |
SAXS
data collected at Bruker Nanostar w Excillum source, Department of Chemistry, iNANO building, Aarhus Uinversity on 2023 Mar 21
|
Dynamics and structural features of the eEF1A1 and eEF1A2 paralogs
Nucleic Acids Research 53(21) (2025)
Novosylna O, Shalak V, Dąbrowska K, Patmanidis I, Lozhko D, Bondarchuk T, Schiøtt B, Pedersen J, Knudsen C, Nissen P, Dadlez M, Negrutskii B
|
| RgGuinier |
2.6 |
nm |
| Dmax |
97.1 |
nm |
| VolumePorod |
60 |
nm3 |
|
|