|
|
|
|
|
| Sample: |
Diguanylate cyclase dimer, 67 kDa Aquella oligotrophica protein
|
| Buffer: |
10 mM Tris, 500 mM NaCl, 2 mM MgCl2, pH: 8
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 May 4
|
Illuminating the inner workings of a natural protein switch: Blue-light sensing in LOV-activated diguanylate cyclases
Science Advances 9(31) (2023)
Vide U, Kasapović D, Fuchs M, Heimböck M, Totaro M, Zenzmaier E, Winkler A
|
| RgGuinier |
3.4 |
nm |
| Dmax |
15.7 |
nm |
| VolumePorod |
104 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Sensor domain-containing diguanylate cyclase dimer, 72 kDa Methylotenera sp. protein
|
| Buffer: |
10 mM Tris, 50 mM NaCl, 2 mM MgCl2, pH: 8
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 May 4
|
Illuminating the inner workings of a natural protein switch: Blue-light sensing in LOV-activated diguanylate cyclases
Science Advances 9(31) (2023)
Vide U, Kasapović D, Fuchs M, Heimböck M, Totaro M, Zenzmaier E, Winkler A
|
| RgGuinier |
2.9 |
nm |
| Dmax |
11.1 |
nm |
| VolumePorod |
115 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Sensor domain-containing diguanylate cyclase dimer, 72 kDa Methylotenera sp. protein
|
| Buffer: |
10 mM Tris, 50 mM NaCl, 2 mM MgCl2, pH: 8
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 May 4
|
Illuminating the inner workings of a natural protein switch: Blue-light sensing in LOV-activated diguanylate cyclases
Science Advances 9(31) (2023)
Vide U, Kasapović D, Fuchs M, Heimböck M, Totaro M, Zenzmaier E, Winkler A
|
| RgGuinier |
3.5 |
nm |
| Dmax |
13.8 |
nm |
| VolumePorod |
123 |
nm3 |
|
|
|
|
|
|
|
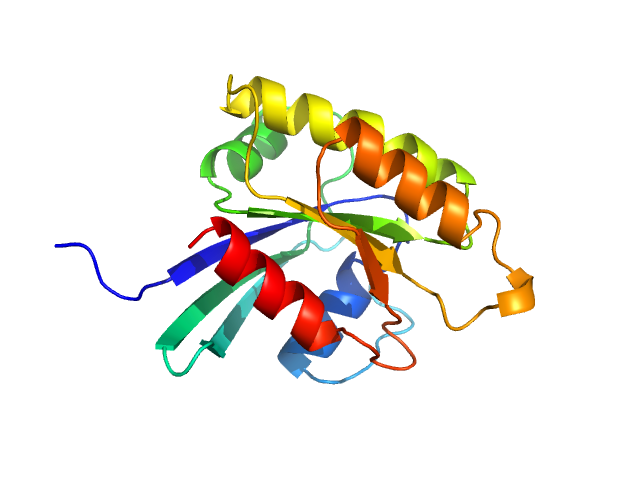
| Sample: |
GTP-binding domain of Ras-like protein 1 monomer, 19 kDa protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 5% glycerol, 3 mM DTT, 5 mM MgCl2, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 21
|
Pathogen-specific structural features of Candida albicans Ras1 activation complex: uncovering new antifungal drug targets.
mBio :e0063823 (2023)
Manso JA, Carabias A, Sárkány Z, de Pereda JM, Pereira PJB, Macedo-Ribeiro S
|
| RgGuinier |
1.6 |
nm |
| Dmax |
4.1 |
nm |
| VolumePorod |
26 |
nm3 |
|
|
|
|
|
|
|
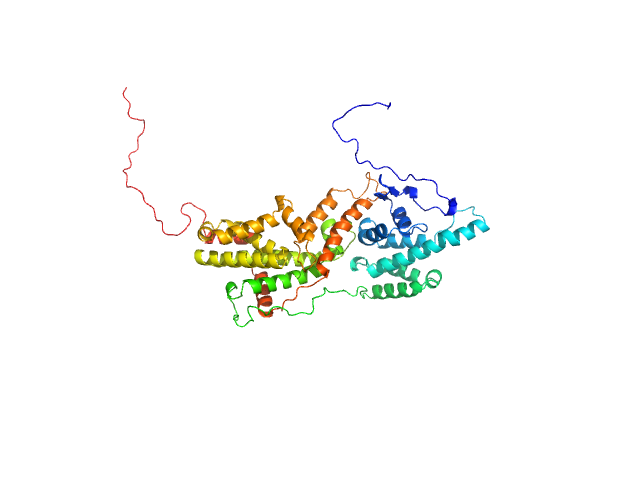
| Sample: |
Ras-like protein 1 monomer, 33 kDa Candida albicans protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 5% glycerol, 3 mM DTT, 5 mM MgCl2, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 21
|
Pathogen-specific structural features of Candida albicans Ras1 activation complex: uncovering new antifungal drug targets.
mBio :e0063823 (2023)
Manso JA, Carabias A, Sárkány Z, de Pereda JM, Pereira PJB, Macedo-Ribeiro S
|
| RgGuinier |
3.6 |
nm |
| Dmax |
15.1 |
nm |
| VolumePorod |
56 |
nm3 |
|
|
|
|
|
|
|
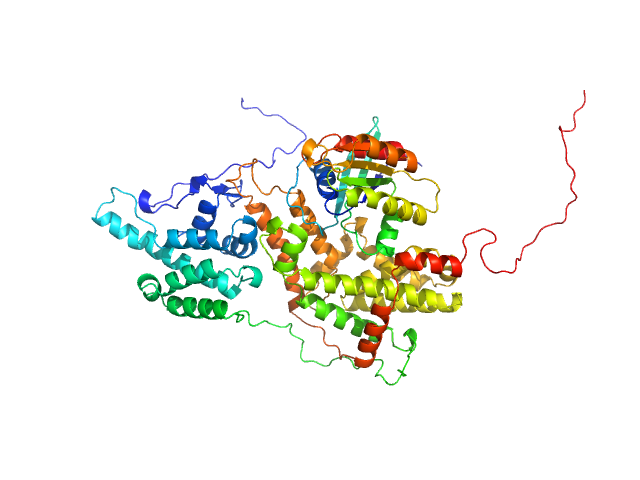
| Sample: |
Cell division control protein 25 monomer, 55 kDa Candida albicans (strain … protein
|
| Buffer: |
20 mM Na-phosphate, 150 mM NaCl, 5% glycerol, 3 mM DTT, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 21
|
Pathogen-specific structural features of Candida albicans Ras1 activation complex: uncovering new antifungal drug targets.
mBio :e0063823 (2023)
Manso JA, Carabias A, Sárkány Z, de Pereda JM, Pereira PJB, Macedo-Ribeiro S
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.8 |
nm |
| VolumePorod |
81 |
nm3 |
|
|
|
|
|
|
|
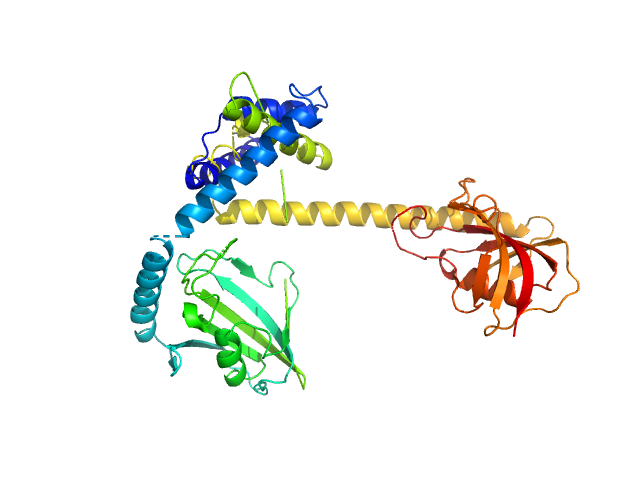
| Sample: |
GTP-binding domain of Ras-like protein 1 monomer, 19 kDa protein
Cell division control protein 25 monomer, 55 kDa Candida albicans (strain … protein
|
| Buffer: |
20 mM Na-phosphate, 150 mM NaCl, 1 mM EDTA, 5% glycerol, 3 mM DTT, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 21
|
Pathogen-specific structural features of Candida albicans Ras1 activation complex: uncovering new antifungal drug targets.
mBio :e0063823 (2023)
Manso JA, Carabias A, Sárkány Z, de Pereda JM, Pereira PJB, Macedo-Ribeiro S
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.8 |
nm |
| VolumePorod |
99 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ras-like protein 1 monomer, 33 kDa Candida albicans protein
Cell division control protein 25 monomer, 55 kDa Candida albicans (strain … protein
|
| Buffer: |
20 mM Na-phosphate, 150 mM NaCl, 1 mM EDTA, 5% glycerol, 3 mM DTT, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 21
|
Pathogen-specific structural features of Candida albicans Ras1 activation complex: uncovering new antifungal drug targets.
mBio :e0063823 (2023)
Manso JA, Carabias A, Sárkány Z, de Pereda JM, Pereira PJB, Macedo-Ribeiro S
|
| RgGuinier |
4.0 |
nm |
| Dmax |
17.7 |
nm |
| VolumePorod |
149 |
nm3 |
|
|
|
|
|
|
|
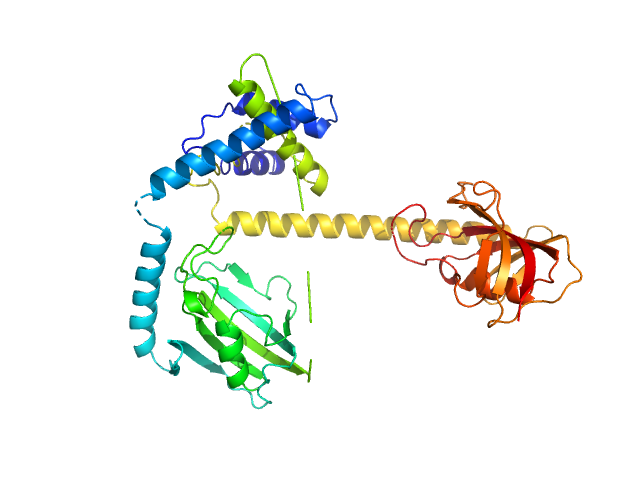
| Sample: |
Outer membrane protein MIP dimer, 46 kDa Legionella pneumophila subsp. … protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 10 mM DTT, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Mar 16
|
Legionella pneumophila macrophage infectivity potentiator protein appendage domains modulate protein dynamics and inhibitor binding
International Journal of Biological Macromolecules :126366 (2023)
Wiedemann C, Whittaker J, Carrillo V, Goretzki B, Dajka M, Tebbe F, Harder J, Krajczy P, Joseph B, Hausch F, Guskov A, Hellmich U
|
| RgGuinier |
3.1 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
66 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Outer membrane protein MIP dimer, 46 kDa Legionella pneumophila subsp. … protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 10 mM DTT, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Mar 16
|
Legionella pneumophila macrophage infectivity potentiator protein appendage domains modulate protein dynamics and inhibitor binding
International Journal of Biological Macromolecules :126366 (2023)
Wiedemann C, Whittaker J, Carrillo V, Goretzki B, Dajka M, Tebbe F, Harder J, Krajczy P, Joseph B, Hausch F, Guskov A, Hellmich U
|
| RgGuinier |
3.1 |
nm |
| Dmax |
10.2 |
nm |
| VolumePorod |
65 |
nm3 |
|
|