|
|
|
|
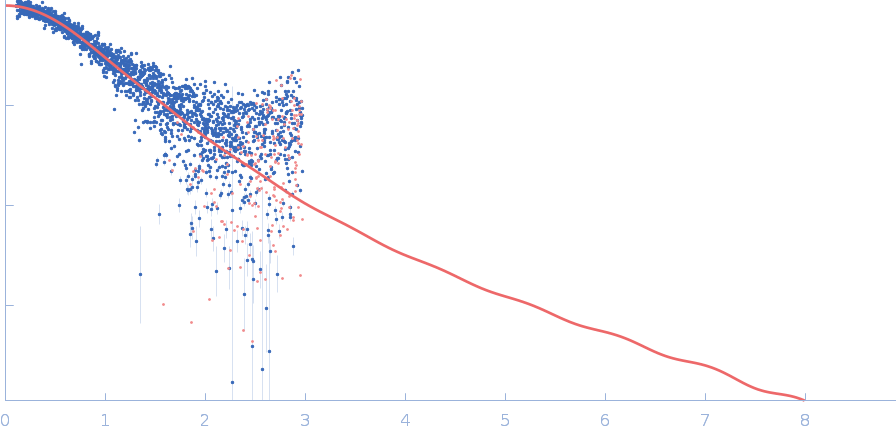
|
| Sample: |
Polyubiquitin-C monomer, 17 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, pH: 7
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 Feb 1
|
Stretching the chains: the destabilizing impact of Cu(2+) and Zn(2+) ions on K48-linked diubiquitin.
Dalton Trans (2023)
Mangini V, Grasso G, Belviso BD, Sciacca MFM, Lanza V, Caliandro R, Milardi D
|
| RgGuinier |
2.0 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
20 |
nm3 |
|
|
|
|
|
|
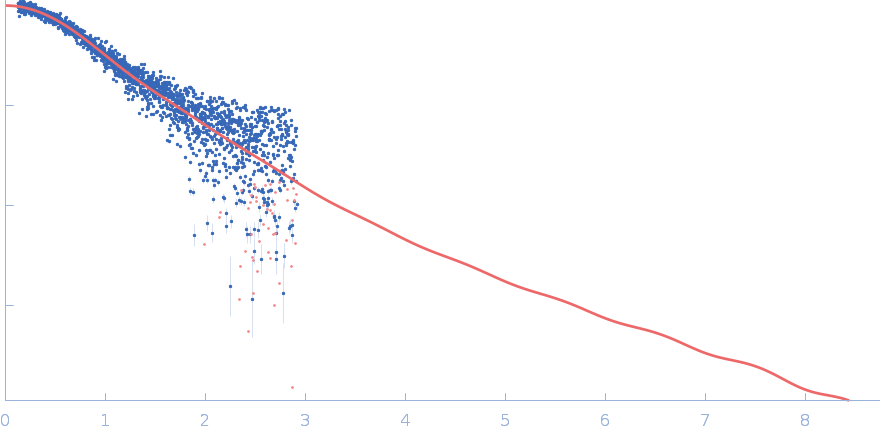
|
| Sample: |
Polyubiquitin-C monomer, 17 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, pH: 7
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 Feb 1
|
Stretching the chains: the destabilizing impact of Cu(2+) and Zn(2+) ions on K48-linked diubiquitin.
Dalton Trans (2023)
Mangini V, Grasso G, Belviso BD, Sciacca MFM, Lanza V, Caliandro R, Milardi D
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.3 |
nm |
| VolumePorod |
20 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Kinesin-like protein KIF3B monomer, 32 kDa Mus musculus protein
Kinesin-associated protein 3 monomer, 81 kDa Mus musculus protein
Kinesin-like protein KIF3A monomer, 28 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris-HCl, 200 mM NaCl, 5% glycerol, 1 mM DTT, pH: 8
|
| Experiment: |
SAXS
data collected at BL-15A2, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2020 Jun 24
|
The two-step cargo recognition mechanism of heterotrimeric kinesin.
EMBO Rep :e56864 (2023)
Jiang X, Ogawa T, Yonezawa K, Shimizu N, Ichinose S, Uchihashi T, Nagaike W, Moriya T, Adachi N, Kawasaki M, Dohmae N, Senda T, Hirokawa N
|
| RgGuinier |
5.8 |
nm |
| Dmax |
27.0 |
nm |
| VolumePorod |
557 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Adenomatous polyposis coli protein (N-terminal ARM domain) monomer, 75 kDa Mus musculus protein
Kinesin-like protein KIF3A monomer, 28 kDa Mus musculus protein
Kinesin-like protein KIF3B monomer, 32 kDa Mus musculus protein
Kinesin-associated protein 3 monomer, 81 kDa Mus musculus protein
|
| Buffer: |
25 mM Tris-HCl, 200 mM NaCl, 1 mM DTT, 5% glycerol, pH: 8
|
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2020 Nov 17
|
The two-step cargo recognition mechanism of heterotrimeric kinesin.
EMBO Rep :e56864 (2023)
Jiang X, Ogawa T, Yonezawa K, Shimizu N, Ichinose S, Uchihashi T, Nagaike W, Moriya T, Adachi N, Kawasaki M, Dohmae N, Senda T, Hirokawa N
|
| RgGuinier |
5.4 |
nm |
| Dmax |
19.5 |
nm |
| VolumePorod |
763 |
nm3 |
|
|
|
|
|
|
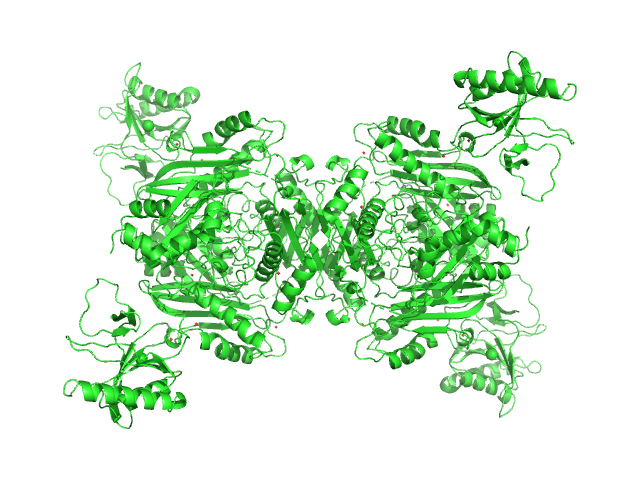
|
| Sample: |
S-adenosylmethionine synthase tetramer, 168 kDa Escherichia coli (strain … protein
S-adenosyl-L-methionine lyase tetramer, 71 kDa Enterobacteria phage T3 protein
|
| Buffer: |
50 mM Tris-HCl, 300 mM NaCl, 5 mM β-mercaptoethanol, 2 % glycerol, pH: 7.5
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2021 Jun 17
|
Phage T3 overcomes the BREX defense through SAM cleavage and inhibition of SAM synthesis by SAM lyase.
Cell Rep 42(8):112972 (2023)
Andriianov A, Trigüis S, Drobiazko A, Sierro N, Ivanov NV, Selmer M, Severinov K, Isaev A
|
| RgGuinier |
4.8 |
nm |
| Dmax |
17.5 |
nm |
| VolumePorod |
325 |
nm3 |
|
|
|
|
|
|
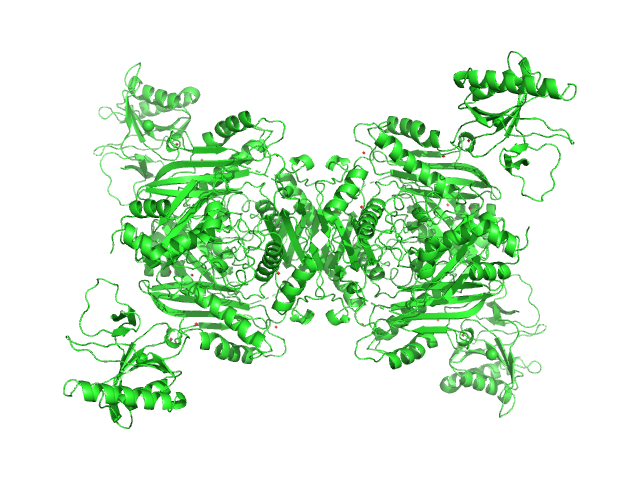
|
| Sample: |
S-adenosylmethionine synthase tetramer, 168 kDa Escherichia coli (strain … protein
S-adenosyl-L-methionine lyase (E68Q/Q94A) tetramer, 71 kDa Enterobacteria phage T3 protein
|
| Buffer: |
50 mM Tris-HCl, 300 mM NaCl, 5 mM β-mercaptoethanol, 2 % glycerol, pH: 7.5
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2021 May 7
|
Phage T3 overcomes the BREX defense through SAM cleavage and inhibition of SAM synthesis by SAM lyase.
Cell Rep 42(8):112972 (2023)
Andriianov A, Trigüis S, Drobiazko A, Sierro N, Ivanov NV, Selmer M, Severinov K, Isaev A
|
| RgGuinier |
4.8 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
326 |
nm3 |
|
|
|
|
|
|
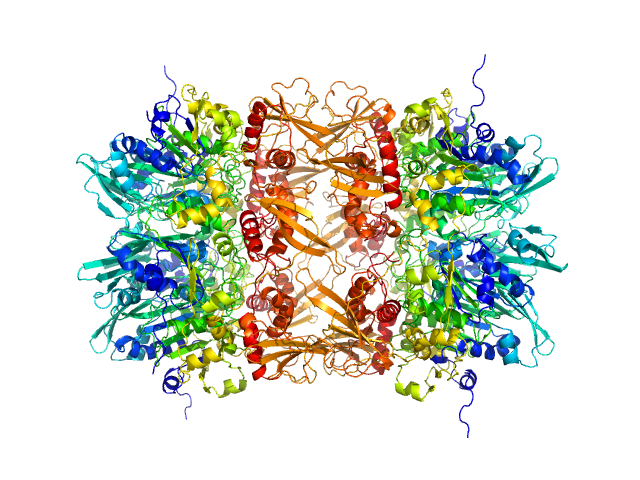
|
| Sample: |
gp11 encapsidation protein decamer, 446 kDa Lactococcus phage Phi28 protein
|
| Buffer: |
100 mM NaCl, 25 mM HEPES, pH: 7.4
|
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, Sealy Center For Structural Biology, UTMB-G on 2015 May 6
|
Lactococcus Phage ASCC Phi28 gp11
Mark White
|
| RgGuinier |
5.3 |
nm |
| Dmax |
16.3 |
nm |
|
|
|
|
|
|
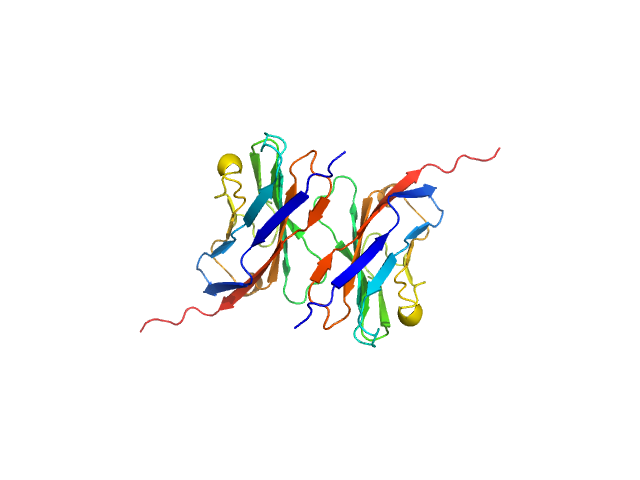
|
| Sample: |
Myelin protein P0 , 15 kDa Homo sapiens protein
|
| Buffer: |
50 mM NaCl, 1 mM EDTA, 20 mM TrisCl, pH: 7.6
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Jul 21
|
Homomeric interactions of the MPZ Ig domain and their relation to Charcot-Marie-Tooth disease
Brain (2023)
Ptak C, Peterson T, Hopkins J, Ahern C, Shy M, Piper R
|
| RgGuinier |
2.9 |
nm |
| Dmax |
17.0 |
nm |
|
|
|
|
|
|
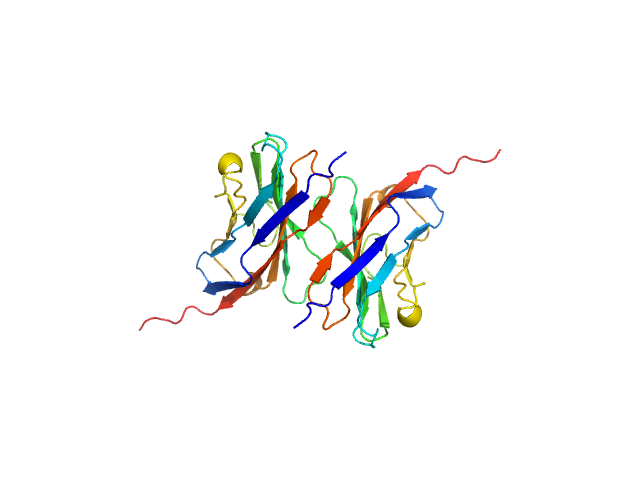
|
| Sample: |
Myelin protein P0 (W53A, R74A, D75R) , 14 kDa Homo sapiens protein
|
| Buffer: |
50 mM NaCl, 1 mM EDTA, 20 mM TrisCl, pH: 7.6
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Jul 14
|
Homomeric interactions of the MPZ Ig domain and their relation to Charcot-Marie-Tooth disease
Brain (2023)
Ptak C, Peterson T, Hopkins J, Ahern C, Shy M, Piper R
|
| RgGuinier |
1.9 |
nm |
| Dmax |
7.0 |
nm |
|
|
|
|
|
|
|
| Sample: |
Diguanylate cyclase dimer, 67 kDa Aquella oligotrophica protein
|
| Buffer: |
10 mM Tris, 500 mM NaCl, 2 mM MgCl2, pH: 8
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 May 4
|
Illuminating the inner workings of a natural protein switch: Blue-light sensing in LOV-activated diguanylate cyclases
Science Advances 9(31) (2023)
Vide U, Kasapović D, Fuchs M, Heimböck M, Totaro M, Zenzmaier E, Winkler A
|
| RgGuinier |
3.4 |
nm |
| Dmax |
13.6 |
nm |
| VolumePorod |
102 |
nm3 |
|
|