|
|
|
|
|
| Sample: |
Protein W monomer, 15 kDa Hendra virus (isolate … protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.2
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 Jun 13
|
Unraveling the molecular grammar and the structural transitions underlying the fibrillation of a viral fibrillogenic domain.
Protein Sci 34(3):e70068 (2025)
Gondelaud F, Leval J, Arora L, Walimbe A, Bignon C, Ptchelkine D, Brocca S, Mukhopadyay S, Longhi S
|
| RgGuinier |
3.4 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
42 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Protein W (artificial PNT3 variant - low-kappa) monomer, 15 kDa Hendra virus (isolate … protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.2
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 Jun 13
|
Unraveling the molecular grammar and the structural transitions underlying the fibrillation of a viral fibrillogenic domain.
Protein Sci 34(3):e70068 (2025)
Gondelaud F, Leval J, Arora L, Walimbe A, Bignon C, Ptchelkine D, Brocca S, Mukhopadyay S, Longhi S
|
| RgGuinier |
3.8 |
nm |
| Dmax |
18.0 |
nm |
| VolumePorod |
48 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Protein W (artificial PNT3 variant - high-kappa) monomer, 15 kDa Hendra virus (isolate … protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.2
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2024 Apr 5
|
Unraveling the molecular grammar and the structural transitions underlying the fibrillation of a viral fibrillogenic domain.
Protein Sci 34(3):e70068 (2025)
Gondelaud F, Leval J, Arora L, Walimbe A, Bignon C, Ptchelkine D, Brocca S, Mukhopadyay S, Longhi S
|
| RgGuinier |
3.1 |
nm |
| Dmax |
14.7 |
nm |
| VolumePorod |
54 |
nm3 |
|
|
|
|
|
|
|
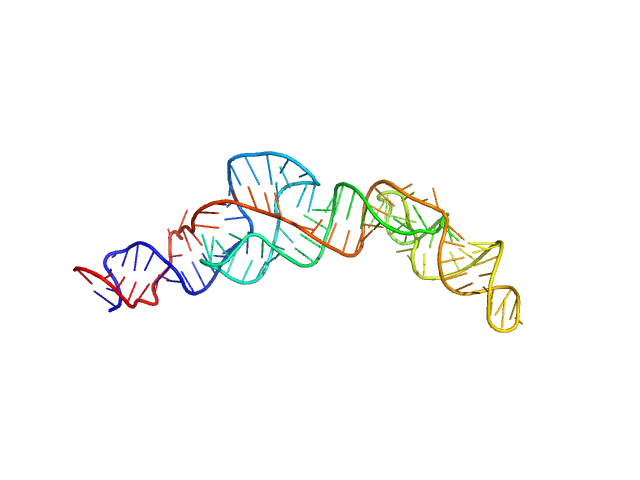
| Sample: |
RNA Device 43 in ligand-free state monomer, 40 kDa synthetic construct RNA
|
| Buffer: |
10 mM Bis-Tris, 100 mM KCl, and 1 mM MgCl2, pH: 6.8
|
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2023 Jul 25
|
Structural investigation of an RNA device that regulates PD-1 expression in mammalian cells.
Nucleic Acids Res 53(5) (2025)
Stagno JR, Deme JC, Dwivedi V, Lee YT, Lee HK, Yu P, Chen SY, Fan L, Degenhardt MFS, Chari R, Young HA, Lea SM, Wang YX
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.2 |
nm |
| VolumePorod |
58 |
nm3 |
|
|
|
|
|
|
|
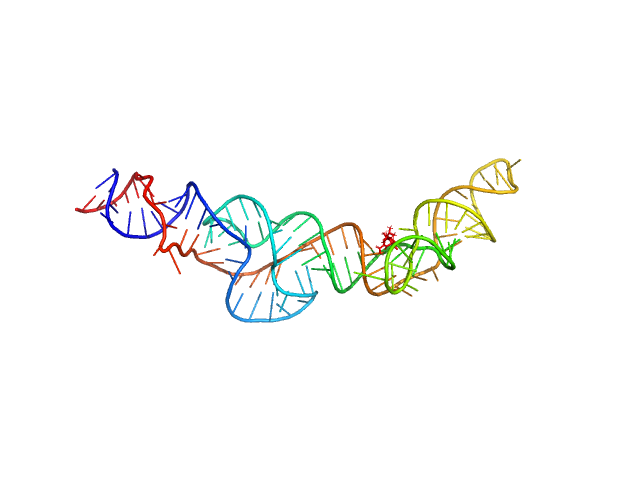
| Sample: |
RNA Device 43 in ligand-bound state monomer, 40 kDa synthetic construct RNA
|
| Buffer: |
10 mM Bis-Tris, 100 mM KCl, and 1 mM MgCl2, pH: 6.8
|
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2023 Jul 25
|
Structural investigation of an RNA device that regulates PD-1 expression in mammalian cells.
Nucleic Acids Res 53(5) (2025)
Stagno JR, Deme JC, Dwivedi V, Lee YT, Lee HK, Yu P, Chen SY, Fan L, Degenhardt MFS, Chari R, Young HA, Lea SM, Wang YX
|
| RgGuinier |
3.3 |
nm |
| Dmax |
12.1 |
nm |
| VolumePorod |
56 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Neurotransmitter-gated ion-channel ligand-binding domain-containing protein pentamer, 342 kDa Desulfofustis sp. PB-SRB1 protein
|
| Buffer: |
20 mM citrate, 150 mM NaCl, 10 mM CaCl2, 0.5 mM deuterated n-Dodecyl-B-D-Maltoside, in D2O, pH: 5
|
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2021 Jun 17
|
Structural characterization of pH-modulated closed and open states in a pentameric ligand-gated ion channel
Marie Lycksell
|
| RgGuinier |
5.1 |
nm |
| Dmax |
17.1 |
nm |
| VolumePorod |
581 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Neurotransmitter-gated ion-channel ligand-binding domain-containing protein pentamer, 342 kDa Desulfofustis sp. PB-SRB1 protein
|
| Buffer: |
20 mM citrate, 150 mM NaCl, 10 mM EDTA, 0.5 mM deuterated n-Dodecyl-β-D-Maltoside, in D2O, pH: 5
|
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2021 Jun 17
|
Structural characterization of pH-modulated closed and open states in a pentameric ligand-gated ion channel
Marie Lycksell
|
| RgGuinier |
5.2 |
nm |
| Dmax |
18.4 |
nm |
| VolumePorod |
558 |
nm3 |
|
|
|
|
|
|
|
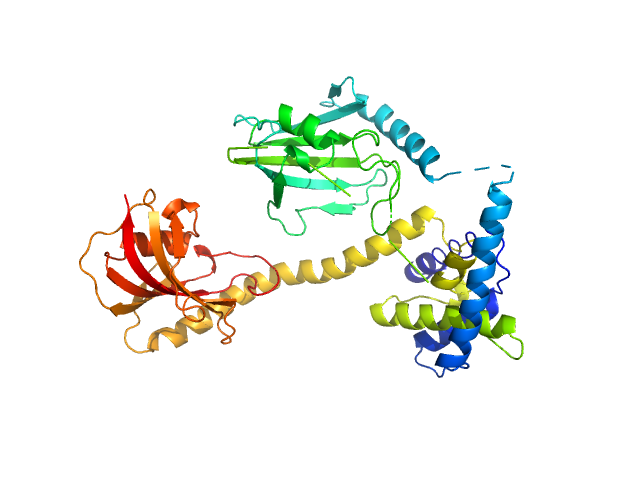
| Sample: |
Outer membrane protein MIP dimer, 46 kDa Legionella pneumophila subsp. … protein
(2S)‐2‐{[(2S)‐1‐[(4‐ fluorophenyl)methanesulfonyl]piperidin‐2‐ yl]formamido}‐4‐methyl‐N‐[(pyridin‐3‐ yl)methyl]pentanamide monomer, 1 kDa
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Nov 22
|
Structure and Dynamics of Macrophage Infectivity Potentiator Proteins from Pathogenic Bacteria and Protozoans Bound to Fluorinated Pipecolic Acid Inhibitors.
J Med Chem (2025)
Pérez Carrillo VH, Whittaker JJ, Wiedemann C, Harder JM, Lohr T, Jamithireddy AK, Dajka M, Goretzki B, Joseph B, Guskov A, Harmer NJ, Holzgrabe U, Hellmich UA
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
61 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Outer membrane protein MIP dimer, 46 kDa Legionella pneumophila subsp. … protein
(2S)‐3‐(4‐fluorophenyl)‐2‐{[(2S)‐1‐[(4‐ fluorophenyl)methanesulfonyl]piperidin‐2‐ yl]formamido}‐N‐[(pyridin‐3‐yl)methyl]propanamide monomer, 1 kDa
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Nov 22
|
Structure and Dynamics of Macrophage Infectivity Potentiator Proteins from Pathogenic Bacteria and Protozoans Bound to Fluorinated Pipecolic Acid Inhibitors.
J Med Chem (2025)
Pérez Carrillo VH, Whittaker JJ, Wiedemann C, Harder JM, Lohr T, Jamithireddy AK, Dajka M, Goretzki B, Joseph B, Guskov A, Harmer NJ, Holzgrabe U, Hellmich UA
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
65 |
nm3 |
|
|
|
|
|
|
|
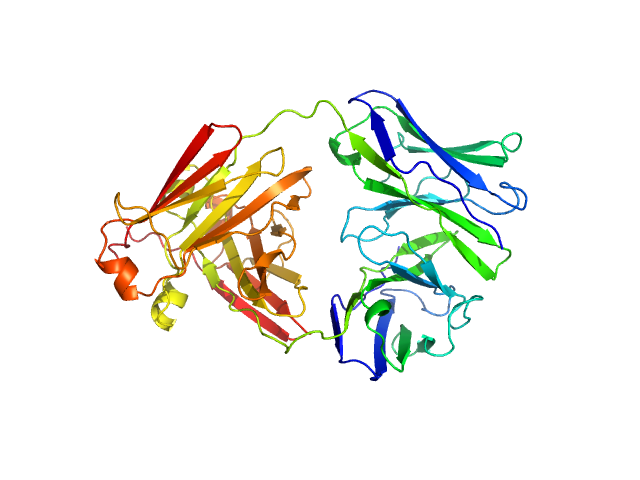
| Sample: |
Immunoglobulin light chain AL55 dimer, 47 kDa Homo sapiens protein
|
| Buffer: |
20 mM TrisHCL, 150 mM NaCl, pH: 8
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Jan 15
|
A conformational fingerprint for amyloidogenic light chains
eLife 13 (2025)
Paissoni C, Puri S, Broggini L, Sriramoju M, Maritan M, Russo R, Speranzini V, Ballabio F, Nuvolone M, Merlini G, Palladini G, Hsu S, Ricagno S, Camilloni C
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
61 |
nm3 |
|
|