|
|
|
|
|
| Sample: |
Early E1A protein monomer, 13 kDa Human adenovirus C … protein
|
| Buffer: |
20 mM sodium phosphate pH 7.0, 200 mM NaCl, 1mM DTT, pH: 7
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Mar 24
|
Conformational buffering underlies functional selection in intrinsically disordered protein regions.
Nat Struct Mol Biol (2022)
González-Foutel NS, Glavina J, Borcherds WM, Safranchik M, Barrera-Vilarmau S, Sagar A, Estaña A, Barozet A, Garrone NA, Fernandez-Ballester G, Blanes-Mira C, Sánchez IE, de Prat-Gay G, Cortés J, Bern...
|
|
|
|
|
|
|
|
| Sample: |
Early E1A protein monomer, 13 kDa Human adenovirus C … protein
|
| Buffer: |
20 mM sodium phosphate pH 7.0, 200 mM NaCl, 1mM DTT, pH: 7
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Mar 24
|
Conformational buffering underlies functional selection in intrinsically disordered protein regions.
Nat Struct Mol Biol (2022)
González-Foutel NS, Glavina J, Borcherds WM, Safranchik M, Barrera-Vilarmau S, Sagar A, Estaña A, Barozet A, Garrone NA, Fernandez-Ballester G, Blanes-Mira C, Sánchez IE, de Prat-Gay G, Cortés J, Bern...
|
|
|
|
|
|
|
|
| Sample: |
Retinoblastoma-associated protein monomer, 41 kDa Homo sapiens protein
Early E1A protein monomer, 13 kDa Human adenovirus C … protein
|
| Buffer: |
20 mM sodium phosphate pH 7.0, 200 mM NaCl, 1mM DTT, pH: 7
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Jul 14
|
Conformational buffering underlies functional selection in intrinsically disordered protein regions.
Nat Struct Mol Biol (2022)
González-Foutel NS, Glavina J, Borcherds WM, Safranchik M, Barrera-Vilarmau S, Sagar A, Estaña A, Barozet A, Garrone NA, Fernandez-Ballester G, Blanes-Mira C, Sánchez IE, de Prat-Gay G, Cortés J, Bern...
|
| RgGuinier |
2.9 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
80 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Retinoblastoma-associated protein monomer, 41 kDa Homo sapiens protein
Early E1A protein monomer, 13 kDa Human adenovirus C … protein
|
| Buffer: |
20 mM sodium phosphate pH 7.0, 200 mM NaCl, 1mM DTT, pH: 7
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Jul 14
|
Conformational buffering underlies functional selection in intrinsically disordered protein regions.
Nat Struct Mol Biol (2022)
González-Foutel NS, Glavina J, Borcherds WM, Safranchik M, Barrera-Vilarmau S, Sagar A, Estaña A, Barozet A, Garrone NA, Fernandez-Ballester G, Blanes-Mira C, Sánchez IE, de Prat-Gay G, Cortés J, Bern...
|
| RgGuinier |
3.0 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
84 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Retinoblastoma-associated protein monomer, 41 kDa Homo sapiens protein
Early E1A protein monomer, 13 kDa Human adenovirus C … protein
|
| Buffer: |
20 mM sodium phosphate pH 7.0, 200 mM NaCl, 1mM DTT, pH: 7
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Jul 14
|
Conformational buffering underlies functional selection in intrinsically disordered protein regions.
Nat Struct Mol Biol (2022)
González-Foutel NS, Glavina J, Borcherds WM, Safranchik M, Barrera-Vilarmau S, Sagar A, Estaña A, Barozet A, Garrone NA, Fernandez-Ballester G, Blanes-Mira C, Sánchez IE, de Prat-Gay G, Cortés J, Bern...
|
| RgGuinier |
3.3 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
93 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Retinoblastoma-associated protein monomer, 41 kDa Homo sapiens protein
Early E1A protein monomer, 13 kDa Human adenovirus C … protein
|
| Buffer: |
20 mM sodium phosphate pH 7.0, 200 mM NaCl, 1mM DTT, pH: 7
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Jul 14
|
Conformational buffering underlies functional selection in intrinsically disordered protein regions.
Nat Struct Mol Biol (2022)
González-Foutel NS, Glavina J, Borcherds WM, Safranchik M, Barrera-Vilarmau S, Sagar A, Estaña A, Barozet A, Garrone NA, Fernandez-Ballester G, Blanes-Mira C, Sánchez IE, de Prat-Gay G, Cortés J, Bern...
|
| RgGuinier |
3.0 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
84 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Early E1A protein monomer, 13 kDa Human adenovirus C … protein
|
| Buffer: |
20 mM sodium phosphate pH 7.0, 200 mM NaCl, 1mM DTT, pH: 7
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Mar 24
|
Conformational buffering underlies functional selection in intrinsically disordered protein regions.
Nat Struct Mol Biol (2022)
González-Foutel NS, Glavina J, Borcherds WM, Safranchik M, Barrera-Vilarmau S, Sagar A, Estaña A, Barozet A, Garrone NA, Fernandez-Ballester G, Blanes-Mira C, Sánchez IE, de Prat-Gay G, Cortés J, Bern...
|
| RgGuinier |
3.6 |
nm |
| Dmax |
17.5 |
nm |
| VolumePorod |
45 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Calmodulin-1 monomer, 17 kDa Homo sapiens protein
|
| Buffer: |
20 mM Hepes, 150 mM NaCl, 2 mM CaCl2, pH: 7.4
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2020 Nov 15
|
Dynamics and structural changes of calmodulin upon interaction with the antagonist calmidazolium.
BMC Biol 20(1):176 (2022)
Léger C, Pitard I, Sadi M, Carvalho N, Brier S, Mechaly A, Raoux-Barbot D, Davi M, Hoos S, Weber P, Vachette P, Durand D, Haouz A, Guijarro JI, Ladant D, Chenal A
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.2 |
nm |
| VolumePorod |
33 |
nm3 |
|
|
|
|
|
|
|
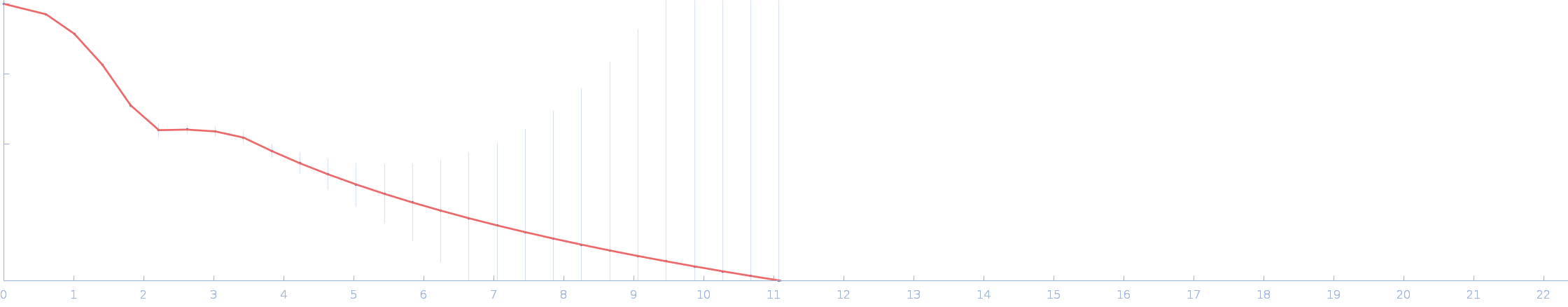
| Sample: |
Calmodulin-1 monomer, 17 kDa Homo sapiens protein
Calmidazolium monomer, 1 kDa
|
| Buffer: |
20 mM Hepes, 150 mM NaCl, 2 mM CaCl2, pH: 7.4
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2020 Nov 15
|
Dynamics and structural changes of calmodulin upon interaction with the antagonist calmidazolium.
BMC Biol 20(1):176 (2022)
Léger C, Pitard I, Sadi M, Carvalho N, Brier S, Mechaly A, Raoux-Barbot D, Davi M, Hoos S, Weber P, Vachette P, Durand D, Haouz A, Guijarro JI, Ladant D, Chenal A
|
| RgGuinier |
1.7 |
nm |
| Dmax |
5.2 |
nm |
| VolumePorod |
30 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Rhoptry kinase family protein tetramer, 143 kDa Toxoplasma gondii (strain … protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, pH: 7.5
|
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2016 Apr 19
|
Divergent kinase WNG1 is regulated by phosphorylation of an atypical activation sub-domain.
Biochem J (2022)
Dewangan PS, Beraki TG, Paiz EA, Appiah Mensah D, Chen Z, Reese ML
|
| RgGuinier |
3.8 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
207 |
nm3 |
|
|