|
|
|
|
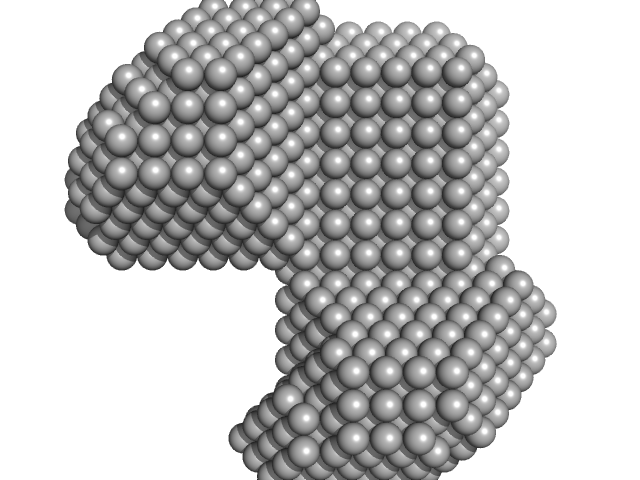
|
| Sample: |
LIM/homeobox protein Lhx3 monomer, 10 kDa Mus musculus protein
|
| Buffer: |
20 mM sodium phosphate monobasic/dibasic, 100 mM NaCl, 1 mM DTT, pH: 7.4
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2018 Nov 2
|
Contrasting DNA-binding behaviour by ISL1 and LHX3 underpins differential gene targeting in neuronal cell specification
Journal of Structural Biology: X :100043 (2020)
Smith N, Wilkinson-White L, Kwan A, Trewhella J, Matthews J
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
18 |
nm3 |
|
|
|
|
|
|
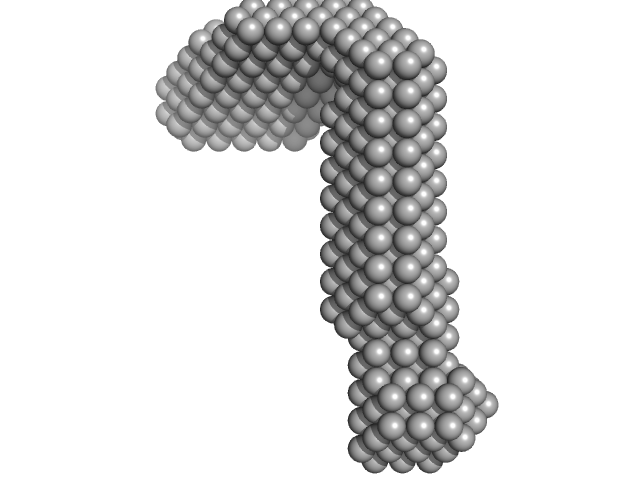
|
| Sample: |
LIM/homeobox protein Lhx3 monomer, 23 kDa Mus musculus protein
Insulin gene enhancer protein ISL-1 monomer, 4 kDa Mus musculus protein
|
| Buffer: |
20 mM sodium phosphate monobasic/dibasic, 100 mM NaCl, 1 mM DTT, pH: 7.4
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2018 Nov 2
|
Contrasting DNA-binding behaviour by ISL1 and LHX3 underpins differential gene targeting in neuronal cell specification
Journal of Structural Biology: X :100043 (2020)
Smith N, Wilkinson-White L, Kwan A, Trewhella J, Matthews J
|
| RgGuinier |
3.3 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
39 |
nm3 |
|
|
|
|
|
|
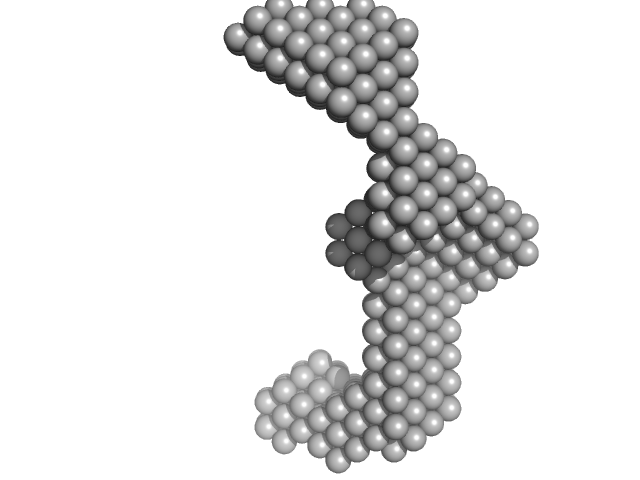
|
| Sample: |
Insulin gene enhancer protein ISL-1 monomer, 12 kDa Mus musculus protein
LIM/homeobox protein Lhx3 monomer, 9 kDa Mus musculus protein
|
| Buffer: |
20 mM sodium phosphate monobasic/dibasic, 100 mM NaCl, 1 mM DTT, pH: 7.4
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2018 Nov 2
|
Contrasting DNA-binding behaviour by ISL1 and LHX3 underpins differential gene targeting in neuronal cell specification
Journal of Structural Biology: X :100043 (2020)
Smith N, Wilkinson-White L, Kwan A, Trewhella J, Matthews J
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.7 |
nm |
| VolumePorod |
40 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Insulin gene enhancer protein ISL-1 monomer, 14 kDa Mus musculus protein
LIM/homeobox protein Lhx3 monomer, 23 kDa Mus musculus protein
|
| Buffer: |
20 mM sodium phosphate monobasic/dibasic, 100 mM NaCl, 1 mM DTT, pH: 7.4
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2018 Nov 2
|
Contrasting DNA-binding behaviour by ISL1 and LHX3 underpins differential gene targeting in neuronal cell specification
Journal of Structural Biology: X :100043 (2020)
Smith N, Wilkinson-White L, Kwan A, Trewhella J, Matthews J
|
| RgGuinier |
3.4 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
76 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
M100 oligonucleotide monomer, 12 kDa DNA
LIM/homeobox protein Lhx3 monomer, 10 kDa Mus musculus protein
|
| Buffer: |
20 mM sodium phosphate monobasic/dibasic, 100 mM NaCl, 1 mM DTT, pH: 7.4
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2018 Nov 2
|
Contrasting DNA-binding behaviour by ISL1 and LHX3 underpins differential gene targeting in neuronal cell specification
Journal of Structural Biology: X :100043 (2020)
Smith N, Wilkinson-White L, Kwan A, Trewhella J, Matthews J
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
27 |
nm3 |
|
|
|
|
|
|
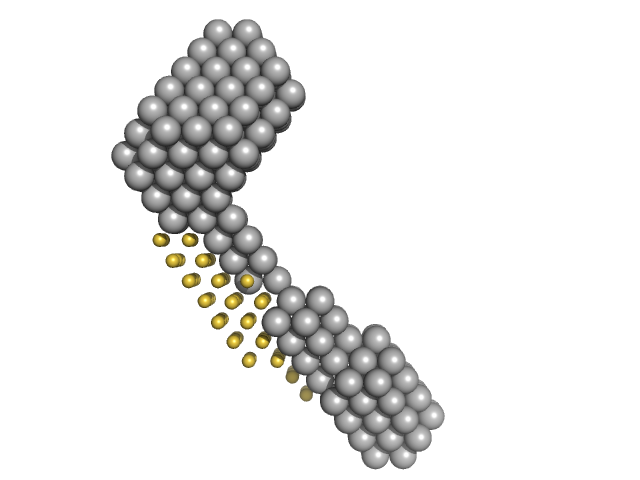
|
| Sample: |
M100 oligonucleotide monomer, 12 kDa DNA
LIM/homeobox protein Lhx3 monomer, 23 kDa Mus musculus protein
Insulin gene enhancer protein ISL-1 monomer, 4 kDa Mus musculus protein
|
| Buffer: |
20 mM sodium phosphate monobasic/dibasic, 100 mM NaCl, 1 mM DTT, pH: 7.4
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2018 Nov 2
|
Contrasting DNA-binding behaviour by ISL1 and LHX3 underpins differential gene targeting in neuronal cell specification
Journal of Structural Biology: X :100043 (2020)
Smith N, Wilkinson-White L, Kwan A, Trewhella J, Matthews J
|
| RgGuinier |
3.6 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
49 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
M100 oligonucleotide monomer, 12 kDa DNA
Insulin gene enhancer protein ISL-1 monomer, 12 kDa Mus musculus protein
LIM/homeobox protein Lhx3 monomer, 9 kDa Mus musculus protein
|
| Buffer: |
20 mM sodium phosphate monobasic/dibasic, 100 mM NaCl, 1 mM DTT, pH: 7.4
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2018 Nov 2
|
Contrasting DNA-binding behaviour by ISL1 and LHX3 underpins differential gene targeting in neuronal cell specification
Journal of Structural Biology: X :100043 (2020)
Smith N, Wilkinson-White L, Kwan A, Trewhella J, Matthews J
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
41 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
M100 oligonucleotide monomer, 12 kDa DNA
Insulin gene enhancer protein ISL-1 monomer, 14 kDa Mus musculus protein
LIM/homeobox protein Lhx3 monomer, 23 kDa Mus musculus protein
|
| Buffer: |
20 mM sodium phosphate monobasic/dibasic, 100 mM NaCl, 1 mM DTT, pH: 7.4
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2018 Nov 2
|
Contrasting DNA-binding behaviour by ISL1 and LHX3 underpins differential gene targeting in neuronal cell specification
Journal of Structural Biology: X :100043 (2020)
Smith N, Wilkinson-White L, Kwan A, Trewhella J, Matthews J
|
| RgGuinier |
3.6 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
70 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
M100 oligonucleotide monomer, 12 kDa DNA
|
| Buffer: |
20 mM sodium phosphate monobasic/dibasic, 100 mM NaCl, 1 mM DTT, pH: 7.4
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2018 Nov 2
|
Contrasting DNA-binding behaviour by ISL1 and LHX3 underpins differential gene targeting in neuronal cell specification
Journal of Structural Biology: X :100043 (2020)
Smith N, Wilkinson-White L, Kwan A, Trewhella J, Matthews J
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.8 |
nm |
| VolumePorod |
15 |
nm3 |
|
|
|
|
|
|
|
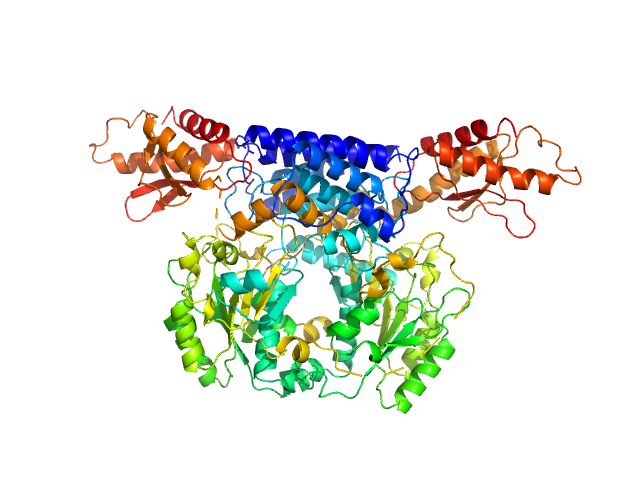
| Sample: |
Cysteine sulfinic acid decarboxylase dimer, 117 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 200 mM NaCl, pH: 7.5
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2018 Feb 4
|
Structure and substrate specificity determinants of the taurine biosynthetic enzyme cysteine sulphinic acid decarboxylase.
J Struct Biol :107674 (2020)
Mahootchi E, Raasakka A, Luan W, Muruganandam G, Loris R, Haavik J, Kursula P
|
| RgGuinier |
3.4 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
198 |
nm3 |
|
|