|
|
|
|
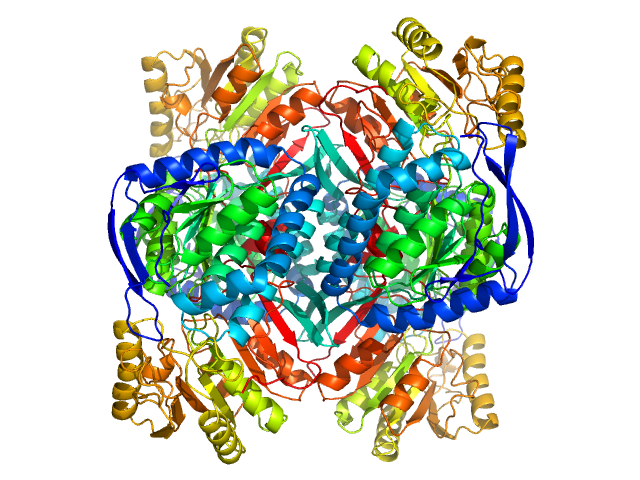
|
| Sample: |
4-trimethylaminobutyraldehyde dehydrogenase tetramer, 215 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 600 mM NaCl, 2% (v/v) glycerol, 1 mM DTT, 1 mM NAD+, pH: 8
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 May 28
|
Inhibition, crystal structures, and in-solution oligomeric structure of aldehyde dehydrogenase 9A1.
Arch Biochem Biophys :108477 (2020)
Wyatt JW, Korasick DA, Qureshi IA, Campbell AC, Gates KS, Tanner JJ
|
| RgGuinier |
3.8 |
nm |
| Dmax |
10.9 |
nm |
| VolumePorod |
252 |
nm3 |
|
|
|
|
|
|
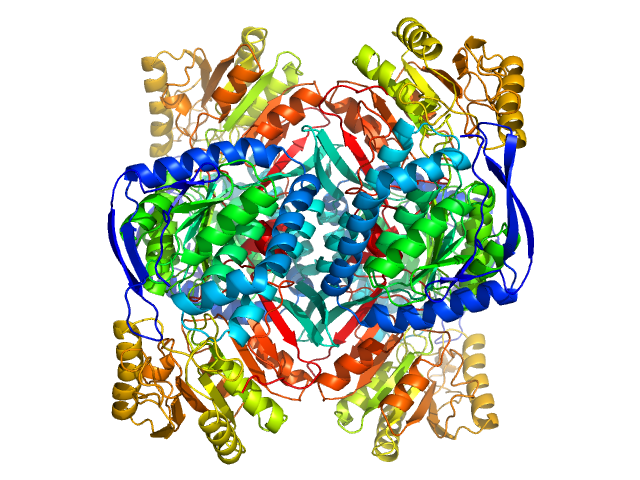
|
| Sample: |
4-trimethylaminobutyraldehyde dehydrogenase tetramer, 215 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 600 mM NaCl, 2% (v/v) glycerol, 1 mM DTT, 1 mM NAD+, pH: 8
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 May 28
|
Inhibition, crystal structures, and in-solution oligomeric structure of aldehyde dehydrogenase 9A1.
Arch Biochem Biophys :108477 (2020)
Wyatt JW, Korasick DA, Qureshi IA, Campbell AC, Gates KS, Tanner JJ
|
| RgGuinier |
3.8 |
nm |
| Dmax |
10.8 |
nm |
| VolumePorod |
240 |
nm3 |
|
|
|
|
|
|
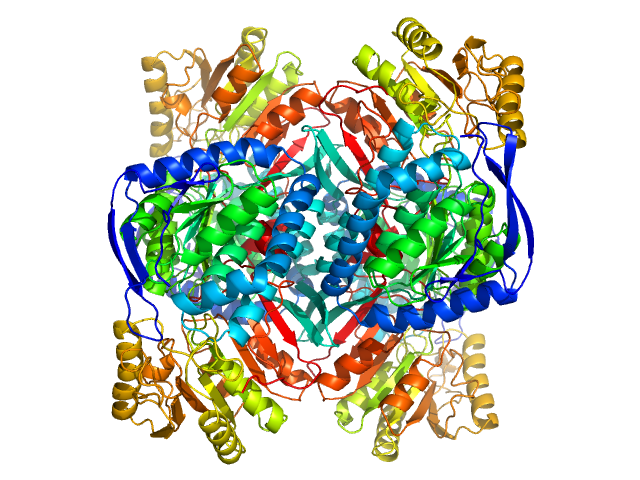
|
| Sample: |
4-trimethylaminobutyraldehyde dehydrogenase tetramer, 215 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 600 mM NaCl, 2% (v/v) glycerol, 1 mM DTT, 1 mM NAD+, pH: 8
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 May 28
|
Inhibition, crystal structures, and in-solution oligomeric structure of aldehyde dehydrogenase 9A1.
Arch Biochem Biophys :108477 (2020)
Wyatt JW, Korasick DA, Qureshi IA, Campbell AC, Gates KS, Tanner JJ
|
| RgGuinier |
3.8 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
240 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
wildtype preQ1 riboswitch in Bacillus subtilis monomer, 11 kDa Bacillus subtilis RNA
|
| Buffer: |
50 mM potassium phosphate, 2 mM MgCl2, 50 mM KCl, pH: 6.5
|
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2018 Mar 23
|
Hierarchical Conformational Dynamics Confers Thermal Adaptability to preQ1 RNA Riboswitches.
J Mol Biol 432(16):4523-4543 (2020)
Gong Z, Yang S, Dong X, Yang QF, Zhu YL, Xiao Y, Tang C
|
| RgGuinier |
2.6 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
20 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
wildtype preQ1 riboswitch in Bacillus subtilis monomer, 11 kDa Bacillus subtilis RNA
|
| Buffer: |
50 mM potassium phosphate, 2 mM MgCl2, 50 mM KCl, pH: 6.5
|
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2018 Mar 23
|
Hierarchical Conformational Dynamics Confers Thermal Adaptability to preQ1 RNA Riboswitches.
J Mol Biol 432(16):4523-4543 (2020)
Gong Z, Yang S, Dong X, Yang QF, Zhu YL, Xiao Y, Tang C
|
| RgGuinier |
2.0 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
16 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
C15 deletion preQ1 riboswitch in Bacillus subtilis monomer, 11 kDa Bacillus subtilis RNA
|
| Buffer: |
50 mM potassium phosphate, 2 mM MgCl2, 50 mM KCl, pH: 6.5
|
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2018 Mar 23
|
Hierarchical Conformational Dynamics Confers Thermal Adaptability to preQ1 RNA Riboswitches.
J Mol Biol 432(16):4523-4543 (2020)
Gong Z, Yang S, Dong X, Yang QF, Zhu YL, Xiao Y, Tang C
|
| RgGuinier |
1.9 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
17 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
C15 deletion preQ1 riboswitch in Bacillus subtilis monomer, 11 kDa Bacillus subtilis RNA
|
| Buffer: |
50 mM potassium phosphate, 2 mM MgCl2, 50 mM KCl, pH: 6.5
|
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2018 Mar 23
|
Hierarchical Conformational Dynamics Confers Thermal Adaptability to preQ1 RNA Riboswitches.
J Mol Biol 432(16):4523-4543 (2020)
Gong Z, Yang S, Dong X, Yang QF, Zhu YL, Xiao Y, Tang C
|
| RgGuinier |
2.0 |
nm |
| Dmax |
8.8 |
nm |
| VolumePorod |
14 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
wildtype preQ1 riboswitch in Thermoanaerobacter tengcongensis monomer, 11 kDa Caldanaerobacter subterraneus subsp. … RNA
|
| Buffer: |
50 mM potassium phosphate, 2 mM MgCl2, 50 mM KCl, pH: 6.5
|
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2018 Mar 23
|
Hierarchical Conformational Dynamics Confers Thermal Adaptability to preQ1 RNA Riboswitches.
J Mol Biol 432(16):4523-4543 (2020)
Gong Z, Yang S, Dong X, Yang QF, Zhu YL, Xiao Y, Tang C
|
| RgGuinier |
1.6 |
nm |
| Dmax |
6.7 |
nm |
| VolumePorod |
12 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
wildtype preQ1 riboswitch in Thermoanaerobacter tengcongensis monomer, 11 kDa Caldanaerobacter subterraneus subsp. … RNA
|
| Buffer: |
50 mM potassium phosphate, 2 mM MgCl2, 50 mM KCl, pH: 6.5
|
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2018 Mar 23
|
Hierarchical Conformational Dynamics Confers Thermal Adaptability to preQ1 RNA Riboswitches.
J Mol Biol 432(16):4523-4543 (2020)
Gong Z, Yang S, Dong X, Yang QF, Zhu YL, Xiao Y, Tang C
|
| RgGuinier |
1.6 |
nm |
| Dmax |
7.2 |
nm |
| VolumePorod |
13 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
wildtype preQ1 riboswitch in Thermoanaerobacter tengcongensis monomer, 11 kDa Caldanaerobacter subterraneus subsp. … RNA
|
| Buffer: |
50 mM potassium phosphate, 2 mM MgCl2, 50 mM KCl, pH: 6.5
|
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2018 Mar 23
|
Hierarchical Conformational Dynamics Confers Thermal Adaptability to preQ1 RNA Riboswitches.
J Mol Biol 432(16):4523-4543 (2020)
Gong Z, Yang S, Dong X, Yang QF, Zhu YL, Xiao Y, Tang C
|
| RgGuinier |
1.7 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
11 |
nm3 |
|
|