|
|
|
|

|
| Sample: |
Bifunctional hemolysin/adenylate cyclase monomer, 57 kDa Bordetella pertussis protein
|
| Buffer: |
10 mM Tris HCl, 150 mM NaCl, 10 mM CaCl₂, pH: 8
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Oct 31
|
Continuous Assembly of β-Roll Structures Is Implicated in the Type I-Dependent Secretion of Large Repeat-in-Toxins (RTX) Proteins.
J Mol Biol (2020)
Motlova L, Klimova N, Fiser R, Sebo P, Bumba L
|
| RgGuinier |
4.0 |
nm |
| Dmax |
13.3 |
nm |
| VolumePorod |
94 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Bifunctional hemolysin/adenylate cyclase monomer, 45 kDa Bordetella pertussis protein
|
| Buffer: |
10 mM Tris HCl, 150 mM NaCl, 10 mM CaCl₂, pH: 8
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Oct 31
|
Continuous Assembly of β-Roll Structures Is Implicated in the Type I-Dependent Secretion of Large Repeat-in-Toxins (RTX) Proteins.
J Mol Biol (2020)
Motlova L, Klimova N, Fiser R, Sebo P, Bumba L
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.1 |
nm |
| VolumePorod |
85 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Bifunctional hemolysin/adenylate cyclase monomer, 32 kDa Bordetella pertussis protein
|
| Buffer: |
10 mM Tris HCl, 150 mM NaCl, 10 mM CaCl₂, pH: 8
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Oct 31
|
Continuous Assembly of β-Roll Structures Is Implicated in the Type I-Dependent Secretion of Large Repeat-in-Toxins (RTX) Proteins.
J Mol Biol (2020)
Motlova L, Klimova N, Fiser R, Sebo P, Bumba L
|
| RgGuinier |
2.7 |
nm |
| Dmax |
8.6 |
nm |
| VolumePorod |
45 |
nm3 |
|
|
|
|
|
|
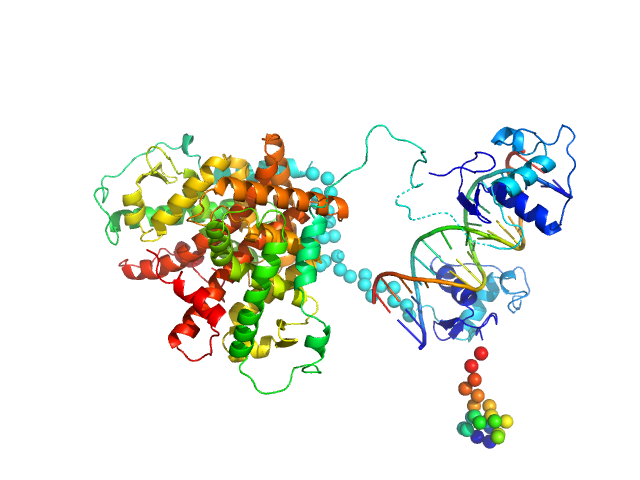
|
| Sample: |
Retinoic acid receptor alpha, RAR monomer, 41 kDa Mus musculus protein
Retinoic acid receptor RXR-alpha monomer, 38 kDa Mus musculus protein
DNA response element HoxB13 DR0 monomer, 10 kDa DNA
|
| Buffer: |
20 mM Tris, pH 8, 150 mM NaCl, 5% v/v glycerol, 1 mM CHAPS, 4 mM MgSO4, 1 mM TCEP, pH: 8
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2014 Jan 19
|
Structural basis for DNA recognition and allosteric control of the retinoic acid receptors RAR–RXR
Nucleic Acids Research (2020)
Osz J, McEwen A, Bourguet M, Przybilla F, Peluso-Iltis C, Poussin-Courmontagne P, Mély Y, Cianférani S, Jeffries C, Svergun D, Rochel N
|
| RgGuinier |
3.8 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
132 |
nm3 |
|
|
|
|
|
|
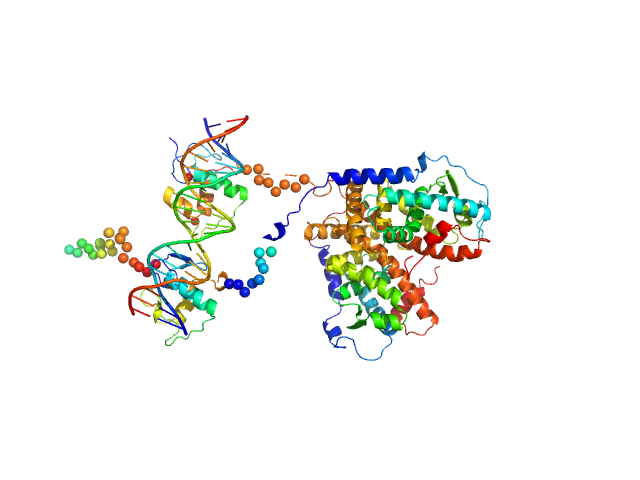
|
| Sample: |
Retinoic acid receptor alpha, RAR monomer, 41 kDa Mus musculus protein
Retinoic acid receptor RXR-alpha monomer, 38 kDa Mus musculus protein
DNA response element F11r DR5 monomer, 13 kDa DNA
|
| Buffer: |
20 mM Tris, pH 8, 150 mM NaCl, 5% v/v glycerol, 1 mM CHAPS, 4 mM MgSO4, 1 mM TCEP, pH: 8
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2014 Jan 19
|
Structural basis for DNA recognition and allosteric control of the retinoic acid receptors RAR–RXR
Nucleic Acids Research (2020)
Osz J, McEwen A, Bourguet M, Przybilla F, Peluso-Iltis C, Poussin-Courmontagne P, Mély Y, Cianférani S, Jeffries C, Svergun D, Rochel N
|
| RgGuinier |
4.0 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
130 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Collagenase ColH (Full-length) monomer, 112 kDa Hathewaya histolytica protein
|
| Buffer: |
10 mM HEPES, 100 mM NaCl, 0.4 mM EGTA, 2.4 mM CaCl2, pH: 7.5
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Dec 12
|
Aromatic residues, Tyr780, His782, Tyr796, Tyr801, located on PKD2 domain of Hathewaya histolytica collagenase, ColH, are involved in the 10-fold enhancement of binding affinity for the enzyme towards...
Perry Caviness
|
| RgGuinier |
4.1 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
198 |
nm3 |
|
|
|
|
|
|
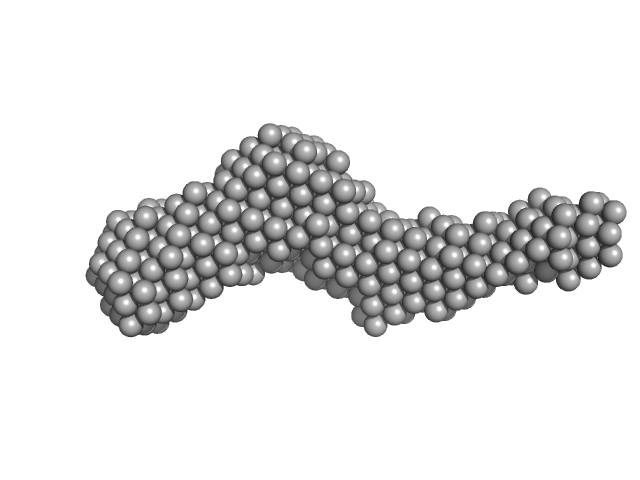
|
| Sample: |
Collagenous Peptide model [(PPG)10] trimer, 9 kDa synthetic construct protein
Collagenase ColH (Polycystic kidney disease domain 2 (PKD2) and Collagen binding domain (CBD)) monomer, 23 kDa Hathewaya histolytica protein
|
| Buffer: |
50 mM HEPES, 100 mM NaCl, 5 mM CaCl2, pH: 7.5
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Dec 12
|
Aromatic residues, Tyr780, His782, Tyr796, Tyr801, located on PKD2 domain of Hathewaya histolytica collagenase, ColH, are involved in the 10-fold enhancement of binding affinity for the enzyme towards...
Perry Caviness
|
| RgGuinier |
3.5 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
41 |
nm3 |
|
|
|
|
|
|
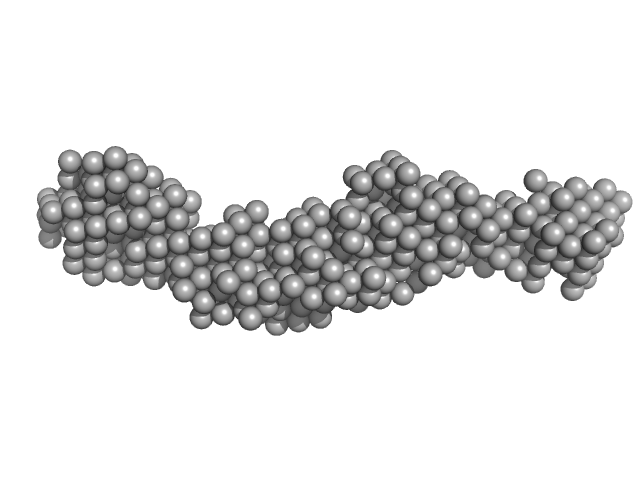
|
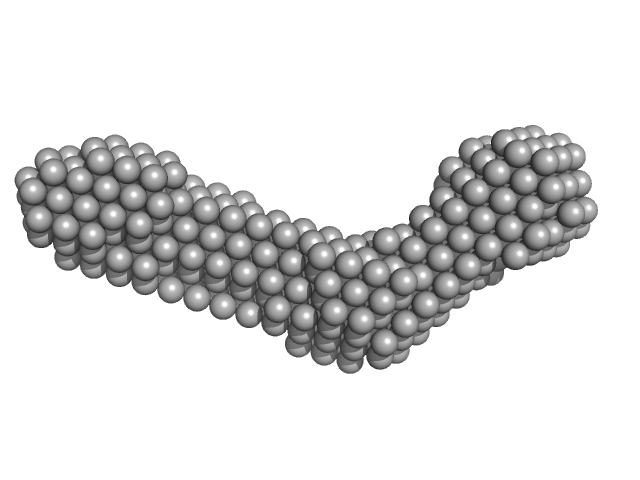
| Sample: |
Collagen like-peptide [GPRG(POG)13] trimer, 11 kDa protein
Collagenase ColH (Polycystic kidney disease 1 (PKD1), Polycystic kidney disease domain 2 (PKD2) and Collagen binding domain (CBD) with Tyr689Ser, Phe712Ser, Tyr780Ser, His782Ser, Tyr796Ser and Tyr801) monomer, 33 kDa Hathewaya histolytica protein
|
| Buffer: |
50 mM HEPES, 100 mM NaCl, 5 mM CaCl2, pH: 7.5
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Mar 5
|
Aromatic residues, Tyr780, His782, Tyr796, Tyr801, located on PKD2 domain of Hathewaya histolytica collagenase, ColH, are involved in the 10-fold enhancement of binding affinity for the enzyme towards...
Perry Caviness
|
| RgGuinier |
4.0 |
nm |
| Dmax |
22.0 |
nm |
| VolumePorod |
61 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Collagenase ColH (Polycystic kidney disease 1 (PKD1), Polycystic kidney disease domain 2 (PKD2) and Collagen binding domain (CBD) with Tyr689Ser, Phe712Ser, Tyr780Ser, His782Ser, Tyr796Ser and Tyr801) monomer, 33 kDa Hathewaya histolytica protein
|
| Buffer: |
50 mM Tris, 1 mM CaCl2, pH: 7.5
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Mar 5
|
Aromatic residues, Tyr780, His782, Tyr796, Tyr801, located on PKD2 domain of Hathewaya histolytica collagenase, ColH, are involved in the 10-fold enhancement of binding affinity for the enzyme towards...
Perry Caviness
|
| RgGuinier |
3.3 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
34 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Collagen like-peptide [GPRG(POG)13] trimer, 11 kDa protein
Collagenase ColH (Polycystic kidney disease 1 (PKD1), Polycystic kidney disease domain 2 (PKD2) and Collagen binding domain (CBD) with Tyr689Ser and Phe712Ser) monomer, 33 kDa Hathewaya histolytica protein
|
| Buffer: |
50 mM HEPES, 100 mM NaCl, 5 mM CaCl2, pH: 7.5
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Dec 12
|
Aromatic residues, Tyr780, His782, Tyr796, Tyr801, located on PKD2 domain of Hathewaya histolytica collagenase, ColH, are involved in the 10-fold enhancement of binding affinity for the enzyme towards...
Perry Caviness
|
| RgGuinier |
4.4 |
nm |
| Dmax |
21.0 |
nm |
| VolumePorod |
79 |
nm3 |
|
|