|
|
|
|

|
| Sample: |
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
36-mer dsRNA monomer, 23 kDa RNA
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM DTT, pH: 7.5
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 May 9
|
Integrative structural analysis of NF45-NF90 heterodimers reveals architectural rearrangements and oligomerization on binding dsRNA.
Nucleic Acids Res 53(6) (2025)
Winterbourne S, Jayachandran U, Zou J, Rappsilber J, Granneman S, Cook AG
|
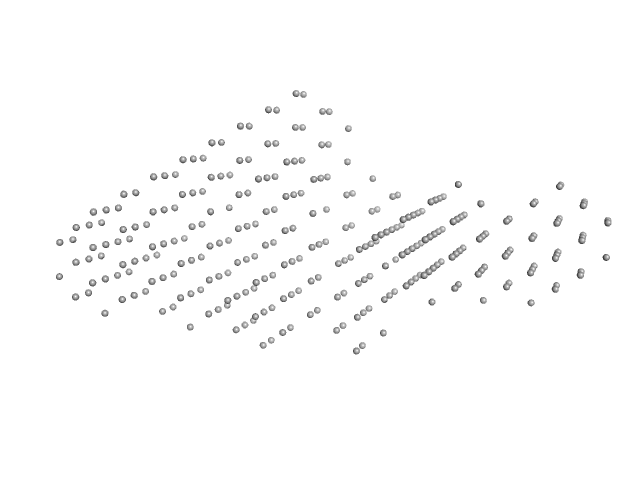
| RgGuinier |
6.4 |
nm |
| Dmax |
22.1 |
nm |
| VolumePorod |
626 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
54-mer dsRNA monomer, 35 kDa RNA
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM DTT, pH: 7.5
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 May 9
|
Integrative structural analysis of NF45-NF90 heterodimers reveals architectural rearrangements and oligomerization on binding dsRNA.
Nucleic Acids Res 53(6) (2025)
Winterbourne S, Jayachandran U, Zou J, Rappsilber J, Granneman S, Cook AG
|
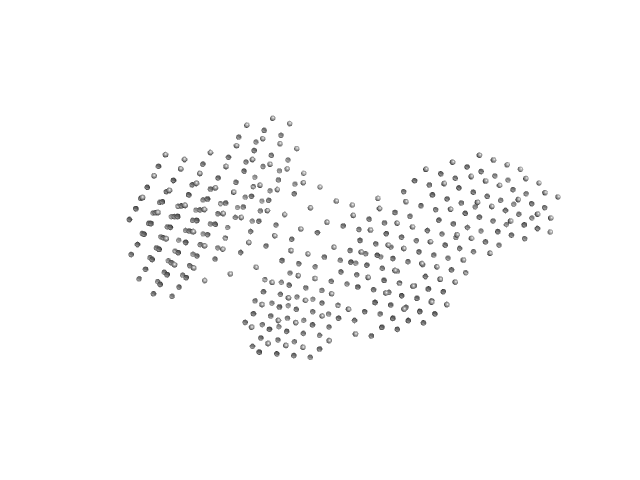
| RgGuinier |
7.1 |
nm |
| Dmax |
25.0 |
nm |
| VolumePorod |
797 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
54-mer dsRNA monomer, 35 kDa RNA
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM DTT, pH: 7.5
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 May 9
|
Integrative structural analysis of NF45-NF90 heterodimers reveals architectural rearrangements and oligomerization on binding dsRNA.
Nucleic Acids Res 53(6) (2025)
Winterbourne S, Jayachandran U, Zou J, Rappsilber J, Granneman S, Cook AG
|
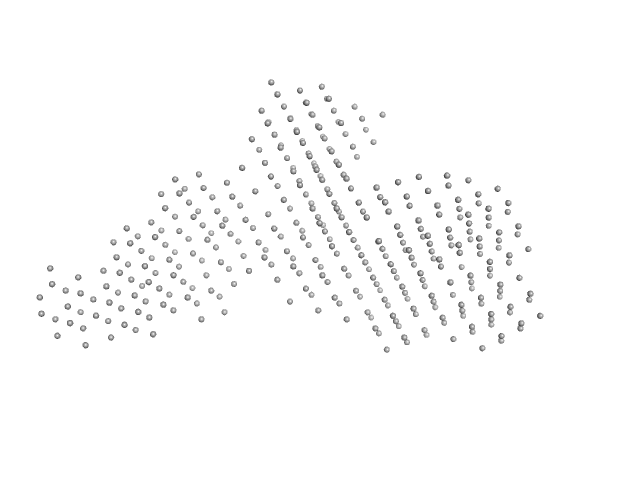
| RgGuinier |
7.7 |
nm |
| Dmax |
26.5 |
nm |
| VolumePorod |
1075 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
54-mer dsRNA monomer, 35 kDa RNA
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM DTT, pH: 7.5
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 May 9
|
Integrative structural analysis of NF45-NF90 heterodimers reveals architectural rearrangements and oligomerization on binding dsRNA.
Nucleic Acids Res 53(6) (2025)
Winterbourne S, Jayachandran U, Zou J, Rappsilber J, Granneman S, Cook AG
|
| RgGuinier |
7.8 |
nm |
| Dmax |
26.1 |
nm |
| VolumePorod |
1050 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Signal transducer and activator of transcription 1-alpha/beta tetramer, 349 kDa Homo sapiens protein
|
| Buffer: |
10 mM HEPES-NaOH, 150 mM NaCl, 3% glycerol, 2 mM DTT, pH: 7.4
|
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2022 Dec 11
|
Structural analysis reveals how tetrameric tyrosine-phosphorylated STAT1 is targeted by the rabies virus P-protein
Science Signaling 18(878) (2025)
Sugiyama A, Minami M, Ugajin K, Inaba-Inoue S, Yabuno N, Takekawa Y, Xiaomei S, Takei S, Sasaki M, Nomai T, Jiang X, Kita S, Maenaka K, Hirose M, Yao M, Gooley P, Moseley G, Sugita Y, Ose T
|
| RgGuinier |
6.2 |
nm |
| Dmax |
21.2 |
nm |
| VolumePorod |
800 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Signal transducer and activator of transcription 1-alpha/beta tetramer, 349 kDa Homo sapiens protein
Phosphoprotein monomer, 33 kDa Rabies virus (strain … protein
|
| Buffer: |
10 mM HEPES-NaOH, 150 mM NaCl, 3% glycerol, 2 mM DTT, pH: 7.4
|
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2022 Dec 11
|
Structural analysis reveals how tetrameric tyrosine-phosphorylated STAT1 is targeted by the rabies virus P-protein
Science Signaling 18(878) (2025)
Sugiyama A, Minami M, Ugajin K, Inaba-Inoue S, Yabuno N, Takekawa Y, Xiaomei S, Takei S, Sasaki M, Nomai T, Jiang X, Kita S, Maenaka K, Hirose M, Yao M, Gooley P, Moseley G, Sugita Y, Ose T
|
| RgGuinier |
6.5 |
nm |
| Dmax |
26.0 |
nm |
| VolumePorod |
857 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Signal transducer and activator of transcription 1-alpha/beta tetramer, 349 kDa Homo sapiens protein
Signal transducer and activator of transcription 1 binding DNA dimer, 11 kDa Homo sapiens DNA
|
| Buffer: |
10 mM HEPES-NaOH, 150 mM NaCl, 3% glycerol, 2 mM DTT, pH: 7.4
|
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2022 Dec 11
|
Structural analysis reveals how tetrameric tyrosine-phosphorylated STAT1 is targeted by the rabies virus P-protein
Science Signaling 18(878) (2025)
Sugiyama A, Minami M, Ugajin K, Inaba-Inoue S, Yabuno N, Takekawa Y, Xiaomei S, Takei S, Sasaki M, Nomai T, Jiang X, Kita S, Maenaka K, Hirose M, Yao M, Gooley P, Moseley G, Sugita Y, Ose T
|
| RgGuinier |
6.1 |
nm |
| Dmax |
19.4 |
nm |
| VolumePorod |
761 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
humanized immunoglobulin G4 monoclonal antibody (lebrikizumab) monomer, 148 kDa Mouse/human protein
|
| Buffer: |
100 mM glycine-HCl, 200 mM NaCl, pH: 2
|
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2024 Nov 19
|
IgG4
and IgG1
undergo common acid‐induced compaction into an alternatively folded state
FEBS Letters (2025)
Imamura H, Honda S
|
| RgGuinier |
4.1 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
238 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
humanized immunoglobulin G1 monoclonal antibody (trastuzumab) monomer, 148 kDa Mouse/human protein
|
| Buffer: |
100 mM glycine-HCl, 200 mM NaCl, pH: 2
|
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2023 May 20
|
IgG4
and IgG1
undergo common acid‐induced compaction into an alternatively folded state
FEBS Letters (2025)
Imamura H, Honda S
|
| RgGuinier |
4.2 |
nm |
| Dmax |
12.9 |
nm |
| VolumePorod |
240 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
humanized immunoglobulin G4 monoclonal antibody (lebrikizumab) monomer, 148 kDa Mouse/human protein
|
| Buffer: |
10 mM sodium phosphate, pH: 7
|
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2024 Nov 19
|
IgG4
and IgG1
undergo common acid‐induced compaction into an alternatively folded state
FEBS Letters (2025)
Imamura H, Honda S
|
| RgGuinier |
4.8 |
nm |
| Dmax |
15.6 |
nm |
| VolumePorod |
211 |
nm3 |
|
|