|
|
|
|
|
| Sample: |
80bp_DNA Forward monomer, 25 kDa Escherichia coli DNA
80bp_DNA Reverse monomer, 25 kDa Escherichia coli DNA
DNA-binding protein HU-alpha , 10 kDa Escherichia coli protein
|
| Buffer: |
10 mM sodium acetate, 150 mM NaCl, pH: 4.5
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Jun 1
|
Nucleoid remodeling during environmental adaptation is regulated by HU-dependent DNA bundling.
Nat Commun 11(1):2905 (2020)
Remesh SG, Verma SC, Chen JH, Ekman AA, Larabell CA, Adhya S, Hammel M
|
|
|
|
|
|
|
|
| Sample: |
80bp_DNA Forward monomer, 25 kDa Escherichia coli DNA
80bp_DNA Reverse monomer, 25 kDa Escherichia coli DNA
DNA-binding protein HU-alpha , 10 kDa Escherichia coli protein
|
| Buffer: |
10 mM sodium acetate, 300 mM NaCl, pH: 4.5
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Jun 1
|
Nucleoid remodeling during environmental adaptation is regulated by HU-dependent DNA bundling.
Nat Commun 11(1):2905 (2020)
Remesh SG, Verma SC, Chen JH, Ekman AA, Larabell CA, Adhya S, Hammel M
|
|
|
|
|
|
|
|
| Sample: |
80bp_DNA Forward monomer, 25 kDa Escherichia coli DNA
80bp_DNA Reverse monomer, 25 kDa Escherichia coli DNA
DNA-binding protein HU-alpha, E34K dimer, 19 kDa Escherichia coli protein
|
| Buffer: |
10 mM sodium acetate, 50 mM NaCl, pH: 4.5
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Nov 2
|
Nucleoid remodeling during environmental adaptation is regulated by HU-dependent DNA bundling.
Nat Commun 11(1):2905 (2020)
Remesh SG, Verma SC, Chen JH, Ekman AA, Larabell CA, Adhya S, Hammel M
|
|
|
|
|
|
|
|
| Sample: |
80bp_DNA Forward monomer, 25 kDa Escherichia coli DNA
80bp_DNA Reverse monomer, 25 kDa Escherichia coli DNA
DNA-binding protein HU-alpha, E34K dimer, 19 kDa Escherichia coli protein
|
| Buffer: |
10 mM Bis-Tris, 50 mM NaCl, pH: 5.5
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Nov 2
|
Nucleoid remodeling during environmental adaptation is regulated by HU-dependent DNA bundling.
Nat Commun 11(1):2905 (2020)
Remesh SG, Verma SC, Chen JH, Ekman AA, Larabell CA, Adhya S, Hammel M
|
|
|
|
|
|
|
|
| Sample: |
80bp_DNA Forward monomer, 25 kDa Escherichia coli DNA
80bp_DNA Reverse monomer, 25 kDa Escherichia coli DNA
DNA-binding protein HU-alpha, E34K , 100 kDa Escherichia coli protein
|
| Buffer: |
10mM Bis-Tris, 50 mM NaCl, pH: 6.5
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Nov 2
|
Nucleoid remodeling during environmental adaptation is regulated by HU-dependent DNA bundling.
Nat Commun 11(1):2905 (2020)
Remesh SG, Verma SC, Chen JH, Ekman AA, Larabell CA, Adhya S, Hammel M
|
| RgGuinier |
7.3 |
nm |
| Dmax |
28.0 |
nm |
| VolumePorod |
330 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
80bp_DNA Forward monomer, 25 kDa Escherichia coli DNA
80bp_DNA Reverse monomer, 25 kDa Escherichia coli DNA
DNA-binding protein HU-alpha, E34K , 100 kDa Escherichia coli protein
|
| Buffer: |
10 mM Bis-Tris, 50 mM NaCl, pH: 7.5
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Nov 2
|
Nucleoid remodeling during environmental adaptation is regulated by HU-dependent DNA bundling.
Nat Commun 11(1):2905 (2020)
Remesh SG, Verma SC, Chen JH, Ekman AA, Larabell CA, Adhya S, Hammel M
|
| RgGuinier |
7.1 |
nm |
| Dmax |
27.5 |
nm |
| VolumePorod |
309 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Aquifex aeolicus McoA metaloxidase ∆328-352 (MCoA∆328-352) monomer, 53 kDa Aquifex aeolicus protein
|
| Buffer: |
50 mM Tris-HCl, 150 mM NaCl, 2 mM TCEP, pH: 7.5
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Jul 13
|
The Methionine-Rich Loop of Multicopper Oxidase McoA follows Open-To-Close Transitions with a Role in Enzyme Catalysis
ACS Catalysis (2020)
Borges P, Brissos V, Hernandez G, Masgrau L, Lucas M, Monza E, Frazão C, Cordeiro T, Martins L
|
| RgGuinier |
2.3 |
nm |
| Dmax |
6.9 |
nm |
| VolumePorod |
77 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Aquifex aeolicus McoA metaloxidase ∆337-346 monomer, 54 kDa Aquifex aeolicus protein
|
| Buffer: |
50 mM Tris-HCl, 150 mM NaCl, 2 mM TCEP, pH: 7.5
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Dec 4
|
The Methionine-Rich Loop of Multicopper Oxidase McoA follows Open-To-Close Transitions with a Role in Enzyme Catalysis
ACS Catalysis (2020)
Borges P, Brissos V, Hernandez G, Masgrau L, Lucas M, Monza E, Frazão C, Cordeiro T, Martins L
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
78 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Aquifex aeolicus McoA metaloxidase monomer, 55 kDa Aquifex aeolicus protein
|
| Buffer: |
50 mM Tris-HCl, 150 mM NaCl, 2 mM TCEP, pH: 7.5
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Apr 15
|
The Methionine-Rich Loop of Multicopper Oxidase McoA follows Open-To-Close Transitions with a Role in Enzyme Catalysis
ACS Catalysis (2020)
Borges P, Brissos V, Hernandez G, Masgrau L, Lucas M, Monza E, Frazão C, Cordeiro T, Martins L
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
79 |
nm3 |
|
|
|
|
|
|
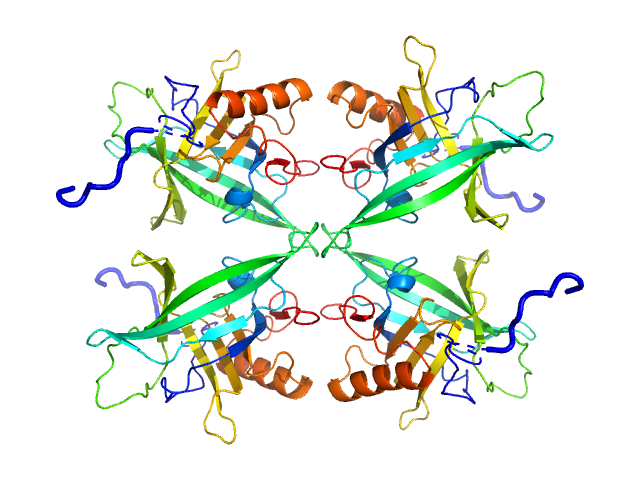
|
| Sample: |
Plasmodium falciparum Lipocalin tetramer, 89 kDa Plasmodium falciparum protein
|
| Buffer: |
20 mM Tris pH7.5, 150 mM NaCl, 5% v/v glycerol, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Apr 8
|
Structure-Based Identification and Functional Characterization of a Lipocalin in the Malaria Parasite Plasmodium falciparum
Cell Reports 31(12):107817 (2020)
Burda P, Crosskey T, Lauk K, Zurborg A, Söhnchen C, Liffner B, Wilcke L, Pietsch E, Strauss J, Jeffries C, Svergun D, Wilson D, Wilmanns M, Gilberger T
|
| RgGuinier |
3.2 |
nm |
| Dmax |
10.3 |
nm |
| VolumePorod |
126 |
nm3 |
|
|