|
|
|
|
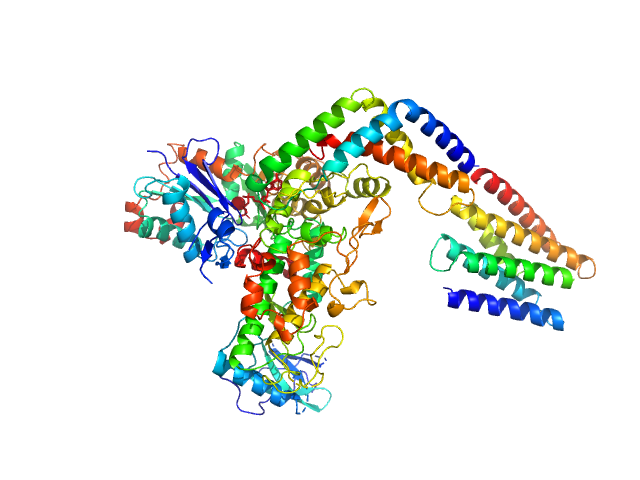
|
| Sample: |
Protein kinase YopO monomer, 63 kDa Yersinia enterocolitica protein
Actin, cytoplasmic 1 monomer, 42 kDa Homo sapiens protein
|
| Buffer: |
10 mM Tris-HCl, 50 mM NaCl, pH: 8
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 May 6
|
Studying Conformational Changes of the Yersinia Type-III-Secretion Effector YopO in Solution by Integrative Structural Biology.
Structure 27(9):1416-1426.e3 (2019)
Peter MF, Tuukkanen AT, Heubach CA, Selsam A, Duthie FG, Svergun DI, Schiemann O, Hagelueken G
|
| RgGuinier |
3.6 |
nm |
| Dmax |
12.3 |
nm |
| VolumePorod |
147 |
nm3 |
|
|
|
|
|
|
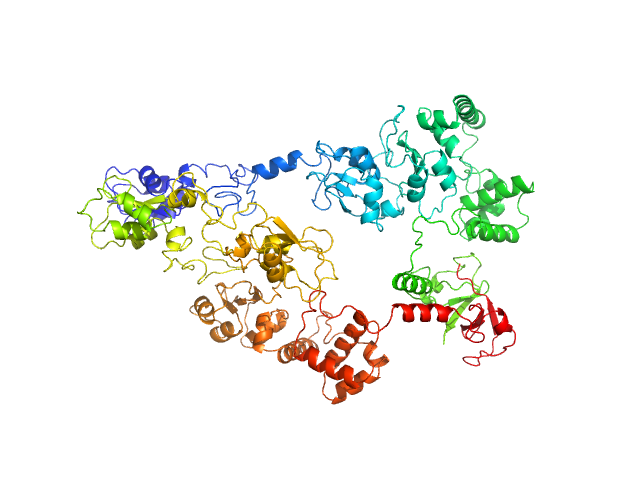
|
| Sample: |
E3 ubiquitin-protein ligase XIAP dimer, 113 kDa Homo sapiens protein
|
| Buffer: |
Xiap buffer, pH: 7.4
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Sep 15
|
Conformational characterization of full-length X-chromosome-linked inhibitor of apoptosis protein (XIAP) through an integrated approach.
IUCrJ 6(Pt 5):948-957 (2019)
Polykretis P, Luchinat E, Bonucci A, Giachetti A, Graewert MA, Svergun DI, Banci L
|
| RgGuinier |
3.9 |
nm |
| Dmax |
12.8 |
nm |
| VolumePorod |
207 |
nm3 |
|
|
|
|
|
|
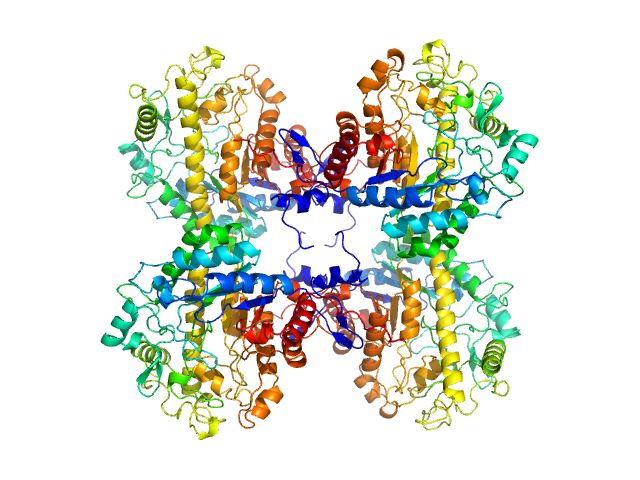
|
| Sample: |
Beta-amylase 2, chloroplastic tetramer, 229 kDa Arabidopsis thaliana protein
|
| Buffer: |
50 mM HEPES, pH: 7.5
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Jun 11
|
Solution structure and assembly of β-amylase2 from Arabidopsis thaliana
(2019)
Chandrasekharan N, Ravenburg C, Roy I, Monroe J, Berndsen C
|
| RgGuinier |
4.2 |
nm |
| Dmax |
12.6 |
nm |
| VolumePorod |
308 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Beta-amylase 2, chloroplastic tetramer, 215 kDa Arabidopsis thaliana protein
|
| Buffer: |
50 mM HEPES, pH: 7.5
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Jun 11
|
Solution structure and assembly of β-amylase2 from Arabidopsis thaliana
(2019)
Chandrasekharan N, Ravenburg C, Roy I, Monroe J, Berndsen C
|
| RgGuinier |
4.4 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
272 |
nm3 |
|
|
|
|
|
|
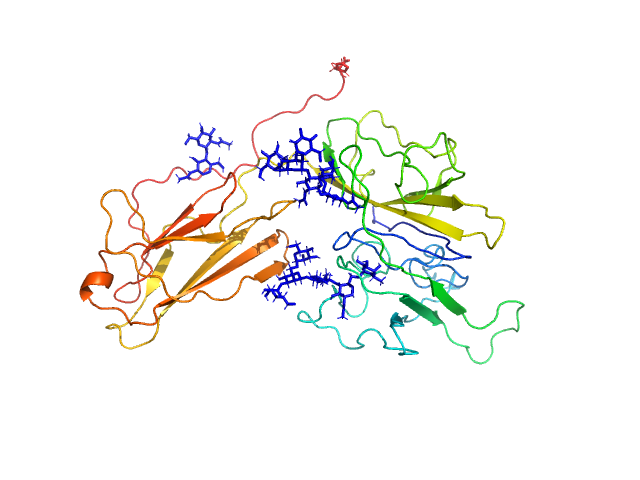
|
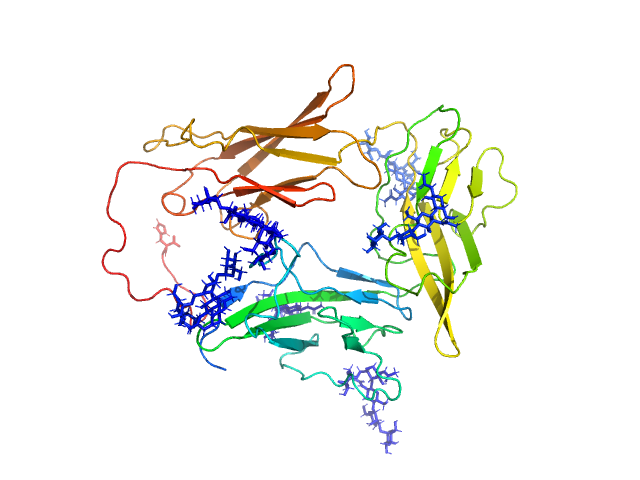
| Sample: |
Interleukin-18 receptor accessory protein monomer, 41 kDa Homo sapiens protein
|
| Buffer: |
10mM HEPES, 150mM NaCl, 3% glycerol, pH: 7.2
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2017 Jul 24
|
Functional Relevance of Interleukin-1 Receptor Inter-domain Flexibility for Cytokine Binding and Signaling.
Structure 27(8):1296-1307.e5 (2019)
Ge J, Remesh SG, Hammel M, Pan S, Mahan AD, Wang S, Wang X
|
| RgGuinier |
3.3 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
70 |
nm3 |
|
|
|
|
|
|
|
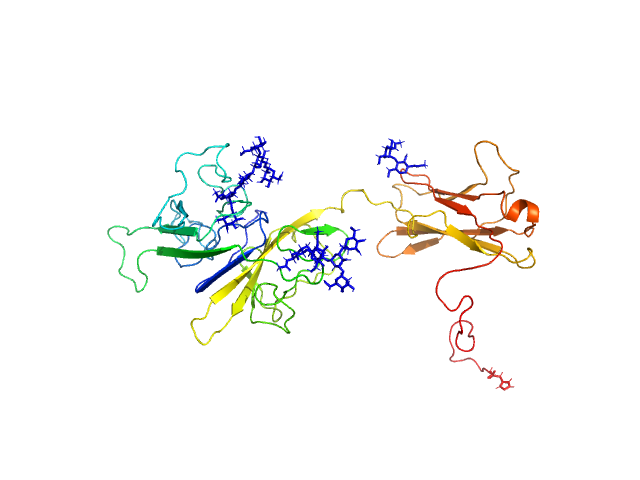
| Sample: |
Interleukin-1 receptor accessory protein ectodomain with RI linker monomer, 41 kDa Homo sapiens protein
|
| Buffer: |
10mM HEPES, 150mM NaCl, 3% glycerol, pH: 7.2
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Aug 11
|
Functional Relevance of Interleukin-1 Receptor Inter-domain Flexibility for Cytokine Binding and Signaling.
Structure 27(8):1296-1307.e5 (2019)
Ge J, Remesh SG, Hammel M, Pan S, Mahan AD, Wang S, Wang X
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.4 |
nm |
| VolumePorod |
76 |
nm3 |
|
|
|
|
|
|
|
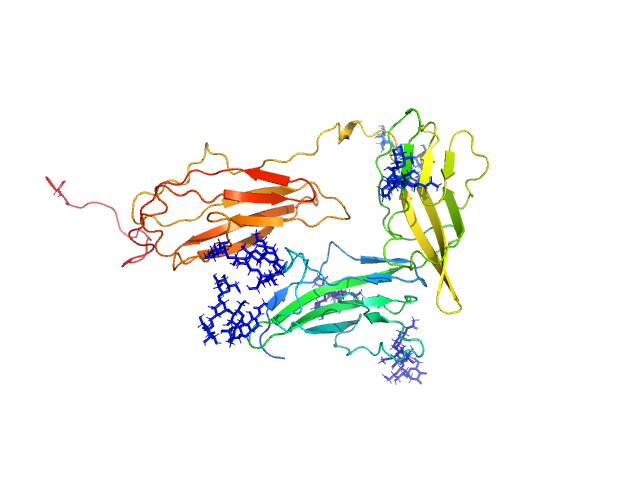
| Sample: |
Interleukin-18 receptor accessory protein ectodomain with Rα linker monomer, 41 kDa Homo sapiens protein
|
| Buffer: |
10mM HEPES, 150mM NaCl, 3% glycerol, pH: 7.2
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2017 Jul 24
|
Functional Relevance of Interleukin-1 Receptor Inter-domain Flexibility for Cytokine Binding and Signaling.
Structure 27(8):1296-1307.e5 (2019)
Ge J, Remesh SG, Hammel M, Pan S, Mahan AD, Wang S, Wang X
|
| RgGuinier |
3.4 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
72 |
nm3 |
|
|
|
|
|
|
|
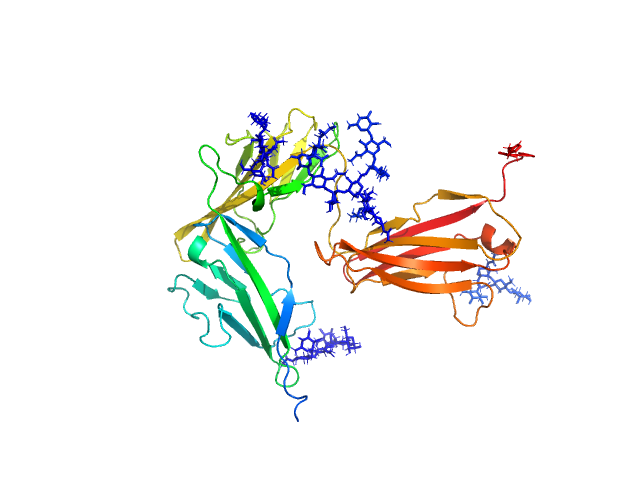
| Sample: |
Interleukin-1 receptor accessory protein ectodomains with ST2 linker monomer, 41 kDa Homo sapiens protein
|
| Buffer: |
10mM HEPES, 150mM NaCl, 3% glycerol, pH: 7.2
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2017 Jul 24
|
Functional Relevance of Interleukin-1 Receptor Inter-domain Flexibility for Cytokine Binding and Signaling.
Structure 27(8):1296-1307.e5 (2019)
Ge J, Remesh SG, Hammel M, Pan S, Mahan AD, Wang S, Wang X
|
| RgGuinier |
3.1 |
nm |
| Dmax |
10.7 |
nm |
| VolumePorod |
76 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Interleukin-1 receptor type 1 monomer, 37 kDa Homo sapiens protein
|
| Buffer: |
10mM HEPES, 150mM NaCl, 3% glycerol, pH: 7.2
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2017 Jul 22
|
Functional Relevance of Interleukin-1 Receptor Inter-domain Flexibility for Cytokine Binding and Signaling.
Structure 27(8):1296-1307.e5 (2019)
Ge J, Remesh SG, Hammel M, Pan S, Mahan AD, Wang S, Wang X
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
63 |
nm3 |
|
|
|
|
|
|
|
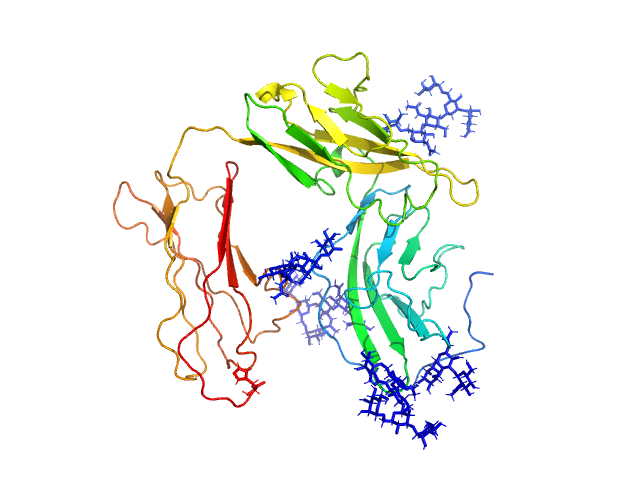
| Sample: |
Interleukin-1 receptor type 2 monomer, 38 kDa Homo sapiens protein
|
| Buffer: |
10mM HEPES, 150mM NaCl, 3% glycerol, pH: 7.2
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2017 Jul 24
|
Functional Relevance of Interleukin-1 Receptor Inter-domain Flexibility for Cytokine Binding and Signaling.
Structure 27(8):1296-1307.e5 (2019)
Ge J, Remesh SG, Hammel M, Pan S, Mahan AD, Wang S, Wang X
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.9 |
nm |
| VolumePorod |
80 |
nm3 |
|
|