|
|
|
|
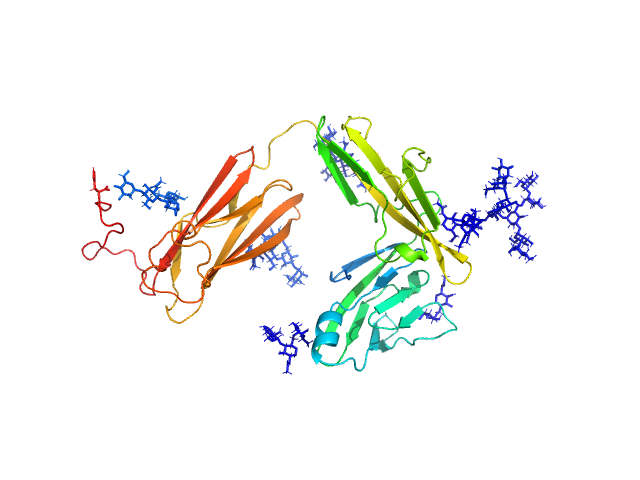
|
| Sample: |
Interleukin-18 receptor 1 monomer, 36 kDa Homo sapiens protein
|
| Buffer: |
10mM HEPES, 150mM NaCl, 3% glycerol, pH: 7.2
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2014 Oct 3
|
Functional Relevance of Interleukin-1 Receptor Inter-domain Flexibility for Cytokine Binding and Signaling.
Structure 27(8):1296-1307.e5 (2019)
Ge J, Remesh SG, Hammel M, Pan S, Mahan AD, Wang S, Wang X
|
| RgGuinier |
3.1 |
nm |
| Dmax |
10.9 |
nm |
| VolumePorod |
77 |
nm3 |
|
|
|
|
|
|
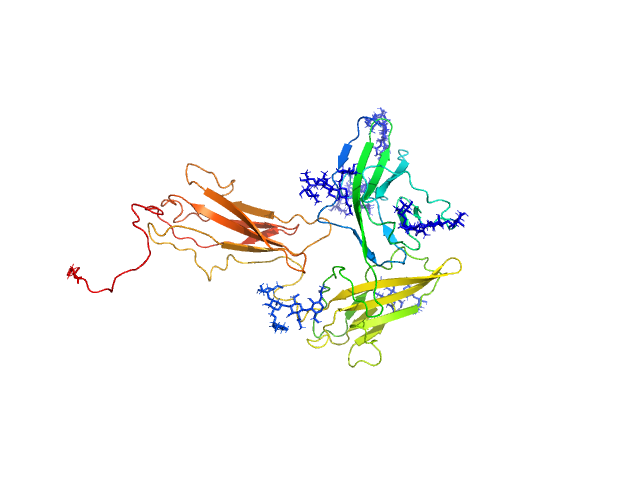
|
| Sample: |
Interleukin-1 receptor accessory protein ectodomains with RII linker monomer, 41 kDa Homo sapiens protein
|
| Buffer: |
10mM HEPES, 150mM NaCl, 3% glycerol, pH: 7.2
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2017 Jul 24
|
Functional Relevance of Interleukin-1 Receptor Inter-domain Flexibility for Cytokine Binding and Signaling.
Structure 27(8):1296-1307.e5 (2019)
Ge J, Remesh SG, Hammel M, Pan S, Mahan AD, Wang S, Wang X
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.7 |
nm |
| VolumePorod |
75 |
nm3 |
|
|
|
|
|
|
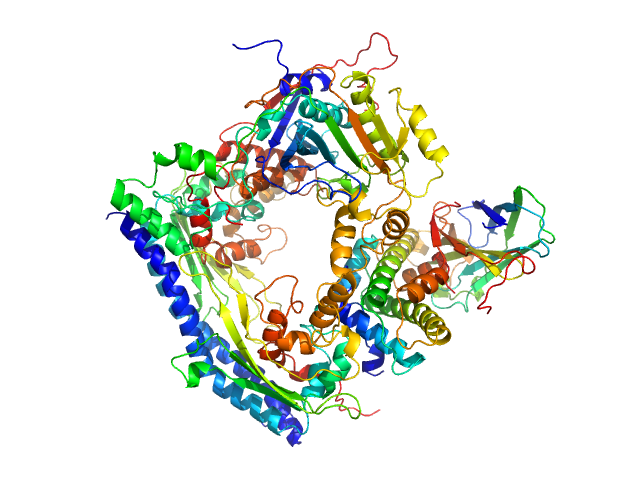
|
| Sample: |
Vacuolar protein sorting-associated protein 75 (1-225 aa) dimer, 53 kDa Saccharomyces cerevisiae protein
Histone acetyltransferase RTT109 monomer, 50 kDa Saccharomyces cerevisiae protein
Histone chaperone ASF1 monomer, 19 kDa protein
Histone H3.2 (35-135 aa) monomer, 12 kDa Xenopus laevis protein
Histone H4 monomer, 11 kDa Xenopus laevis protein
|
| Buffer: |
50 mM citrate, 150 mM NaCl, 5 mM BME, 100% D2O, pH: 6.5
|
| Experiment: |
SANS
data collected at KWS1, FRM2 on 2017 Mar 3
|
Histone chaperone exploits intrinsic disorder to switch acetylation specificity.
Nat Commun 10(1):3435 (2019)
Danilenko N, Lercher L, Kirkpatrick J, Gabel F, Codutti L, Carlomagno T
|
| RgGuinier |
3.5 |
nm |
| Dmax |
11.8 |
nm |
|
|
|
|
|
|
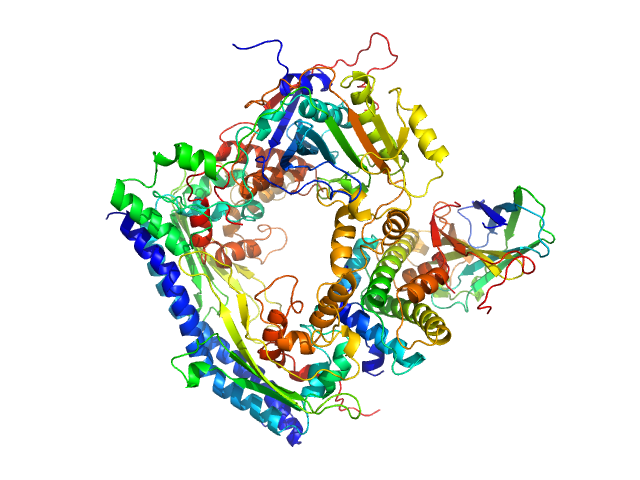
|
| Sample: |
Vacuolar protein sorting-associated protein 75 (1-225 aa) dimer, 53 kDa Saccharomyces cerevisiae protein
Histone acetyltransferase RTT109 monomer, 50 kDa Saccharomyces cerevisiae protein
Histone chaperone ASF1 monomer, 19 kDa protein
Histone H3.2 (35-135 aa) monomer, 12 kDa Xenopus laevis protein
Histone H4 monomer, 11 kDa Xenopus laevis protein
|
| Buffer: |
50 mM citrate, 150 mM NaCl, 5 mM BME, 100% D2O, pH: 6.5
|
| Experiment: |
SANS
data collected at KWS1, FRM2 on 2017 Mar 4
|
Histone chaperone exploits intrinsic disorder to switch acetylation specificity.
Nat Commun 10(1):3435 (2019)
Danilenko N, Lercher L, Kirkpatrick J, Gabel F, Codutti L, Carlomagno T
|
|
|
|
|
|
|
|
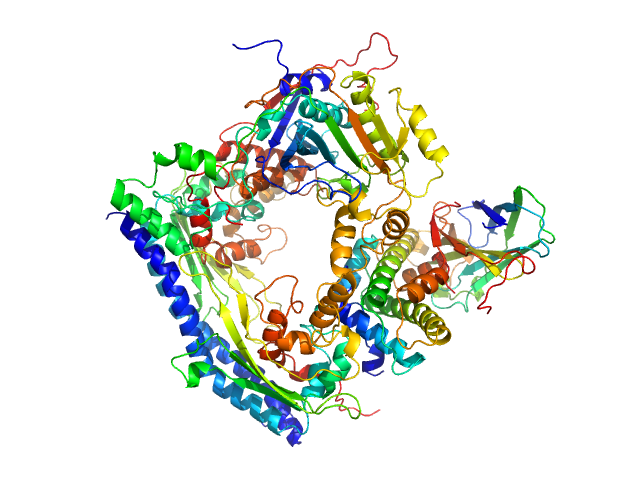
| Sample: |
Vacuolar protein sorting-associated protein 75 (1-225 aa) dimer, 53 kDa Saccharomyces cerevisiae protein
Histone acetyltransferase RTT109 monomer, 50 kDa Saccharomyces cerevisiae protein
Histone chaperone ASF1 monomer, 19 kDa protein
Histone H3.2 (35-135 aa) monomer, 12 kDa Xenopus laevis protein
Histone H4 monomer, 11 kDa Xenopus laevis protein
|
| Buffer: |
50 mM citrate, 150 mM NaCl, 5 mM BME, 100% D2O, pH: 6.5
|
| Experiment: |
SANS
data collected at KWS1, FRM2 on 2017 Mar 4
|
Histone chaperone exploits intrinsic disorder to switch acetylation specificity.
Nat Commun 10(1):3435 (2019)
Danilenko N, Lercher L, Kirkpatrick J, Gabel F, Codutti L, Carlomagno T
|
| RgGuinier |
3.3 |
nm |
| Dmax |
10.5 |
nm |
|
|
|
|
|
|
|
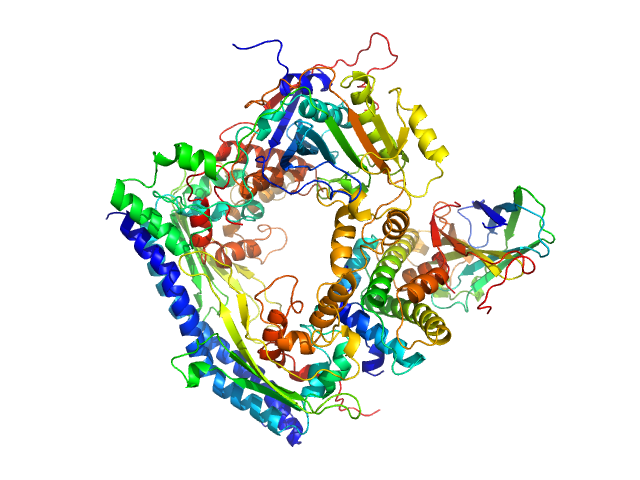
| Sample: |
Vacuolar protein sorting-associated protein 75 (1-225 aa) dimer, 53 kDa Saccharomyces cerevisiae protein
Histone acetyltransferase RTT109 monomer, 50 kDa Saccharomyces cerevisiae protein
Histone chaperone ASF1 monomer, 19 kDa protein
Histone H3.2 (35-135 aa) monomer, 12 kDa Xenopus laevis protein
Histone H4 monomer, 11 kDa Xenopus laevis protein
|
| Buffer: |
50 mM citrate, 150 mM NaCl, 5 mM BME, 100% D2O, pH: 6.5
|
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2018 May 29
|
Histone chaperone exploits intrinsic disorder to switch acetylation specificity.
Nat Commun 10(1):3435 (2019)
Danilenko N, Lercher L, Kirkpatrick J, Gabel F, Codutti L, Carlomagno T
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.5 |
nm |
|
|
|
|
|
|
|
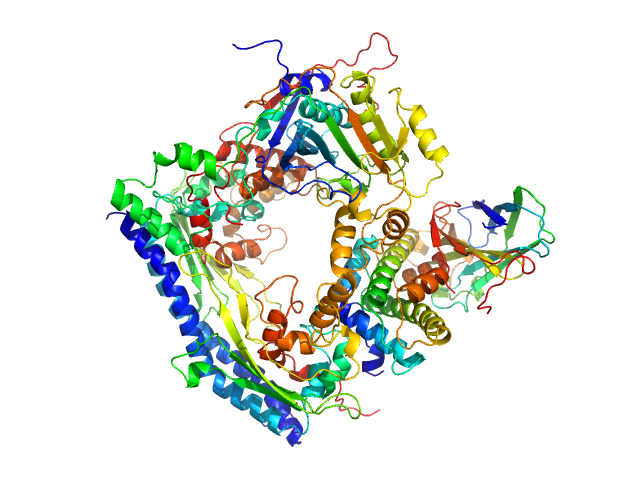
| Sample: |
Vacuolar protein sorting-associated protein 75 (1-225 aa) dimer, 53 kDa Saccharomyces cerevisiae protein
Histone acetyltransferase RTT109 monomer, 50 kDa Saccharomyces cerevisiae protein
Histone chaperone ASF1 monomer, 19 kDa protein
Histone H3.2 (35-135 aa) monomer, 12 kDa Xenopus laevis protein
Histone H4 monomer, 11 kDa Xenopus laevis protein
|
| Buffer: |
50 mM citrate, 150 mM NaCl, 5 mM BME, 42% D2O, pH: 6.5
|
| Experiment: |
SANS
data collected at KWS1, FRM2 on 2017 Mar 5
|
Histone chaperone exploits intrinsic disorder to switch acetylation specificity.
Nat Commun 10(1):3435 (2019)
Danilenko N, Lercher L, Kirkpatrick J, Gabel F, Codutti L, Carlomagno T
|
| RgGuinier |
3.5 |
nm |
| Dmax |
11.0 |
nm |
|
|
|
|
|
|
|
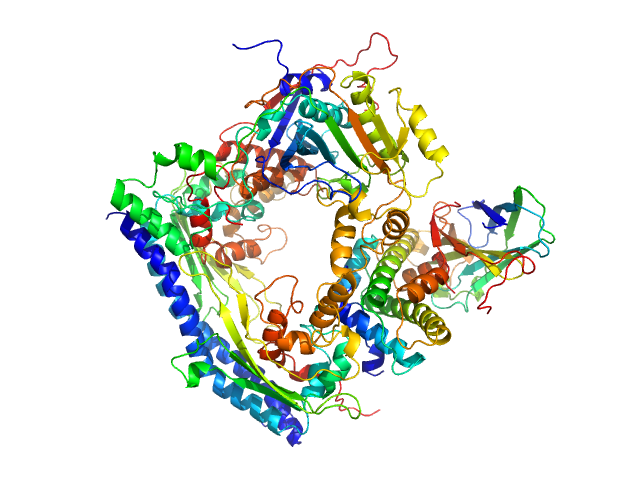
| Sample: |
Vacuolar protein sorting-associated protein 75 (1-225 aa) dimer, 53 kDa Saccharomyces cerevisiae protein
Histone acetyltransferase RTT109 monomer, 50 kDa Saccharomyces cerevisiae protein
Histone chaperone ASF1 monomer, 19 kDa protein
Histone H3.2 (35-135 aa) monomer, 12 kDa Xenopus laevis protein
Histone H4 monomer, 11 kDa Xenopus laevis protein
|
| Buffer: |
50 mM citrate, 150 mM NaCl, 5 mM BME, 42% D2O, pH: 6.5
|
| Experiment: |
SANS
data collected at KWS1, FRM2 on 2017 Mar 5
|
Histone chaperone exploits intrinsic disorder to switch acetylation specificity.
Nat Commun 10(1):3435 (2019)
Danilenko N, Lercher L, Kirkpatrick J, Gabel F, Codutti L, Carlomagno T
|
| RgGuinier |
3.1 |
nm |
| Dmax |
10.5 |
nm |
|
|
|
|
|
|
|
| Sample: |
Vacuolar protein sorting-associated protein 75 (1-225 aa) dimer, 53 kDa Saccharomyces cerevisiae protein
Histone acetyltransferase RTT109 monomer, 50 kDa Saccharomyces cerevisiae protein
Histone chaperone ASF1 monomer, 19 kDa protein
Histone H3.2 (35-135 aa) monomer, 12 kDa Xenopus laevis protein
Histone H4 monomer, 11 kDa Xenopus laevis protein
|
| Buffer: |
50 mM citrate, 150 mM NaCl, 5 mM BME, 100% D2O, pH: 6.5
|
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2016 Nov 14
|
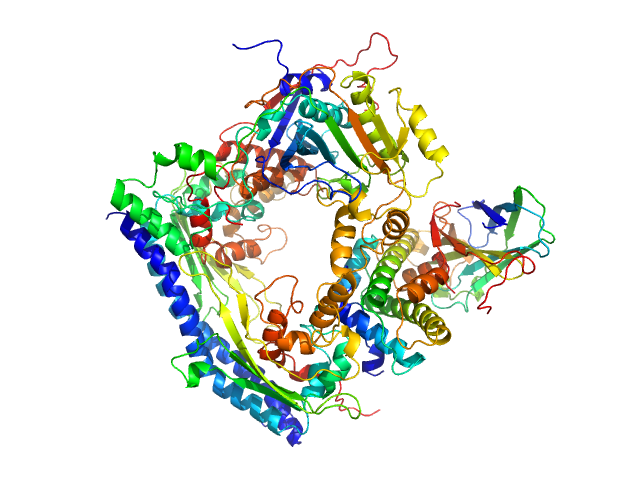
Histone chaperone exploits intrinsic disorder to switch acetylation specificity (Asf1-H3:H4-Rtt109-Vps75 protein complex, data for docking block selections)
Nat Commun 10(1):3435 (2019)
Danilenko N, Lercher L, Kirkpatrick J, Gabel F, Codutti L, Carlomagno T
|
| RgGuinier |
1.9 |
nm |
| Dmax |
5.5 |
nm |
|
|
|
|
|
|
|
| Sample: |
Vacuolar protein sorting-associated protein 75 (1-225 aa) dimer, 53 kDa Saccharomyces cerevisiae protein
Histone acetyltransferase RTT109 monomer, 50 kDa Saccharomyces cerevisiae protein
Histone chaperone ASF1 monomer, 19 kDa protein
Histone H4 monomer, 11 kDa Xenopus laevis protein
Histone H3 full-length monomer, 15 kDa Xenopus laevis protein
|
| Buffer: |
50 mM citrate, 150 mM NaCl, 5 mM BME, 42% D2O, pH: 6.5
|
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2016 Nov 14
|
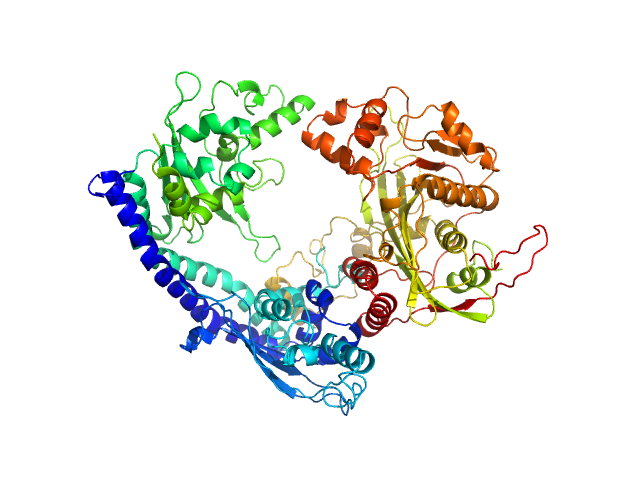
Histone chaperone exploits intrinsic disorder to switch acetylation specificity (Asf1-H3:H4-Rtt109-Vps75 protein complex, data for docking block selections)
Nat Commun 10(1):3435 (2019)
Danilenko N, Lercher L, Kirkpatrick J, Gabel F, Codutti L, Carlomagno T
|
| RgGuinier |
3.4 |
nm |
| Dmax |
10.5 |
nm |
|
|