|
|
|
|

|
| Sample: |
Phox Homology (PX) - C2 domains of human Phosphatidylinositol 4-phosphate 3-kinase C2 domain-containing subunit alpha monomer, 33 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris 200 mM NaCl 5% Glycerol 0.5 mM TCEP, pH: 8.5
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Oct 20
|
Molecular Basis for Membrane Recruitment by the PX and C2 Domains of Class II Phosphoinositide 3-Kinase-C2α.
Structure (2018)
Chen KE, Tillu VA, Chandra M, Collins BM
|
| RgGuinier |
2.6 |
nm |
| Dmax |
9.3 |
nm |
| VolumePorod |
43 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Phox Homology (PX) - C2 domains of human Phosphatidylinositol 4-phosphate 3-kinase C2 domain-containing subunit alpha monomer, 33 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris 200 mM NaCl 5% Glycerol 0.5 mM TCEP 4 mM InsP6, pH: 8.5
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Oct 20
|
Molecular Basis for Membrane Recruitment by the PX and C2 Domains of Class II Phosphoinositide 3-Kinase-C2α.
Structure (2018)
Chen KE, Tillu VA, Chandra M, Collins BM
|
| RgGuinier |
2.6 |
nm |
| Dmax |
9.3 |
nm |
| VolumePorod |
48 |
nm3 |
|
|
|
|
|
|
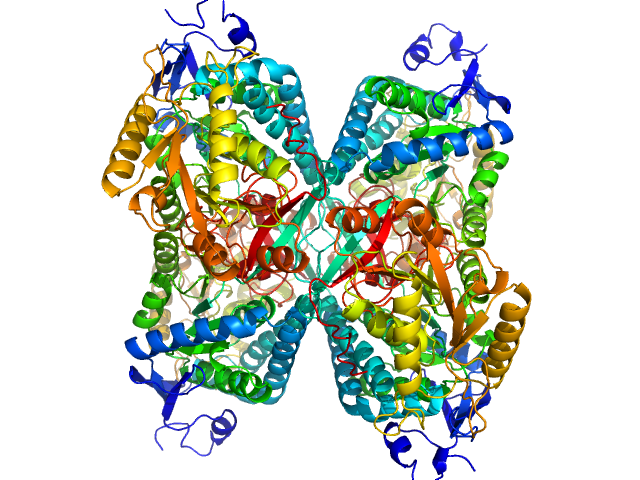
|
| Sample: |
Alpha-aminoadipic semialdehyde dehydrogenase tetramer, 222 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris, 50 mM NaCl, 0.5 mM DTT, 5% (v/v) glycerol, pH: 7.8
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2018 Feb 22
|
NAD+ Promotes Assembly of the Active Tetramer of Aldehyde Dehydrogenase 7A1.
FEBS Lett (2018)
Korasick DA, White TA, Chakravarthy S, Tanner JJ
|
| RgGuinier |
3.5 |
nm |
| VolumePorod |
212 |
nm3 |
|
|
|
|
|
|
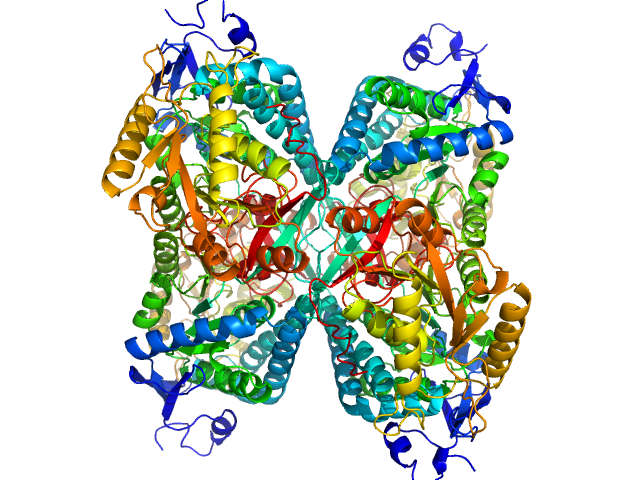
|
| Sample: |
Alpha-aminoadipic semialdehyde dehydrogenase tetramer, 222 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris, 50 mM NaCl, 0.5 mM DTT, 5% (v/v) glycerol, pH: 7.8
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2018 Feb 22
|
NAD+ Promotes Assembly of the Active Tetramer of Aldehyde Dehydrogenase 7A1.
FEBS Lett (2018)
Korasick DA, White TA, Chakravarthy S, Tanner JJ
|
| RgGuinier |
3.7 |
nm |
| VolumePorod |
229 |
nm3 |
|
|
|
|
|
|
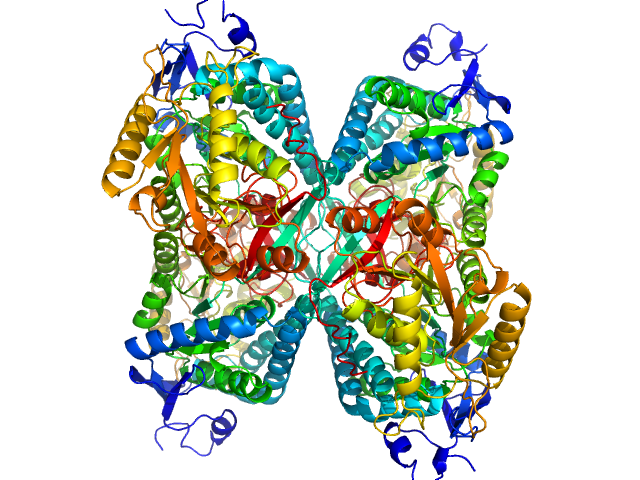
|
| Sample: |
Alpha-aminoadipic semialdehyde dehydrogenase tetramer, 222 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris, 50 mM NaCl, 0.5 mM DTT, 5% (v/v) glycerol, pH: 7.8
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2018 Feb 22
|
NAD+ Promotes Assembly of the Active Tetramer of Aldehyde Dehydrogenase 7A1.
FEBS Lett (2018)
Korasick DA, White TA, Chakravarthy S, Tanner JJ
|
| RgGuinier |
3.7 |
nm |
| VolumePorod |
238 |
nm3 |
|
|
|
|
|
|
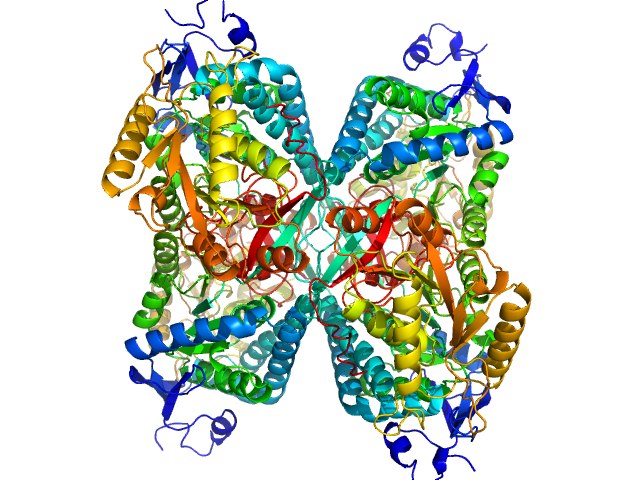
|
| Sample: |
Alpha-aminoadipic semialdehyde dehydrogenase tetramer, 222 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris, 50 mM NaCl, 0.5 mM DTT, 5% (v/v) glycerol, 1 mM NAD, pH: 7.8
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2018 Feb 22
|
NAD+ Promotes Assembly of the Active Tetramer of Aldehyde Dehydrogenase 7A1.
FEBS Lett (2018)
Korasick DA, White TA, Chakravarthy S, Tanner JJ
|
| RgGuinier |
3.7 |
nm |
| VolumePorod |
277 |
nm3 |
|
|
|
|
|
|
|
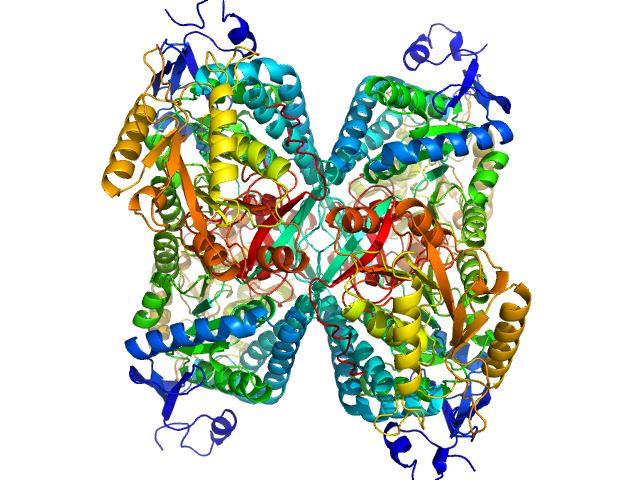
| Sample: |
Alpha-aminoadipic semialdehyde dehydrogenase tetramer, 222 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris, 50 mM NaCl, 0.5 mM DTT, 5% (v/v) glycerol, 1 mM NAD, pH: 7.8
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2018 Feb 22
|
NAD+ Promotes Assembly of the Active Tetramer of Aldehyde Dehydrogenase 7A1.
FEBS Lett (2018)
Korasick DA, White TA, Chakravarthy S, Tanner JJ
|
| RgGuinier |
3.8 |
nm |
| VolumePorod |
275 |
nm3 |
|
|
|
|
|
|
|
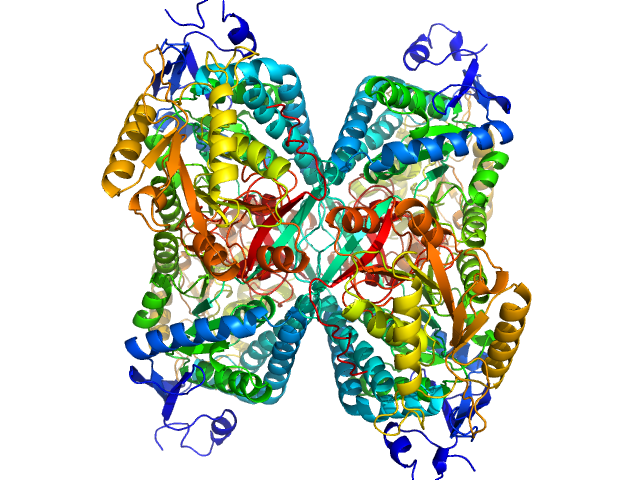
| Sample: |
Alpha-aminoadipic semialdehyde dehydrogenase tetramer, 222 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris, 50 mM NaCl, 0.5 mM DTT, 5% (v/v) glycerol, 1 mM NAD, pH: 7.8
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2018 Feb 22
|
NAD+ Promotes Assembly of the Active Tetramer of Aldehyde Dehydrogenase 7A1.
FEBS Lett (2018)
Korasick DA, White TA, Chakravarthy S, Tanner JJ
|
| RgGuinier |
3.8 |
nm |
| VolumePorod |
277 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
p-hydroxyphenylacetate 3-hydroxylase, reductase component dimer, 71 kDa Acinetobacter baumannii protein
|
| Buffer: |
50 mM MOPS, 0.5 mM EDTA, 1 mM DTT, and 5% glycerol, pH: 7
|
| Experiment: |
SAXS
data collected at BL1.3W, Synchrotron Light Research Institute (SLRI) on 2017 Mar 7
|
Crystal structure of the flavin reductase of Acinetobacter baumannii p-hydroxyphenylacetate 3-hydroxylase (HPAH) and identification of amino acid residues underlying its regulation by aromatic ligands...
Arch Biochem Biophys 653:24-38 (2018)
Yuenyao A, Petchyam N, Kamonsutthipaijit N, Chaiyen P, Pakotiprapha D
|
| RgGuinier |
2.4 |
nm |
| Dmax |
6.9 |
nm |
| VolumePorod |
94 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
p-hydroxyphenylacetate 3-hydroxylase, reductase component dimer, 71 kDa Acinetobacter baumannii protein
|
| Buffer: |
50 mM MOPS, 0.5 mM EDTA, 1 mM DTT, and 5% glycerol, pH: 7
|
| Experiment: |
SAXS
data collected at BL1.3W, Synchrotron Light Research Institute (SLRI) on 2017 Mar 7
|
Crystal structure of the flavin reductase of Acinetobacter baumannii p-hydroxyphenylacetate 3-hydroxylase (HPAH) and identification of amino acid residues underlying its regulation by aromatic ligands...
Arch Biochem Biophys 653:24-38 (2018)
Yuenyao A, Petchyam N, Kamonsutthipaijit N, Chaiyen P, Pakotiprapha D
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.1 |
nm |
| VolumePorod |
95 |
nm3 |
|
|