|
|
|
|

|
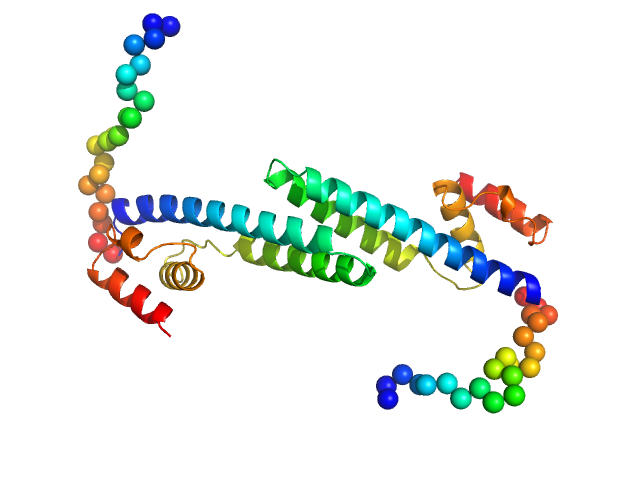
| Sample: |
Calbindin monomer, 30 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150mM NaCl, pH: 7.8
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Feb 28
|
The X-ray structure of human calbindin-D28K: an improved model.
Acta Crystallogr D Struct Biol 74(Pt 10):1008-1014 (2018)
Noble JW, Almalki R, Roe SM, Wagner A, Duman R, Atack JR
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
46 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Fluorescence recovery protein dimer, 28 kDa Synechocystis sp. PCC … protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 3% glycerol, pH: 7.6
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Sep 1
|
OCP-FRP protein complex topologies suggest a mechanism for controlling high light tolerance in cyanobacteria.
Nat Commun 9(1):3869 (2018)
Sluchanko NN, Slonimskiy YB, Shirshin EA, Moldenhauer M, Friedrich T, Maksimov EG
|
| RgGuinier |
2.9 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
43 |
nm3 |
|
|
|
|
|
|
|
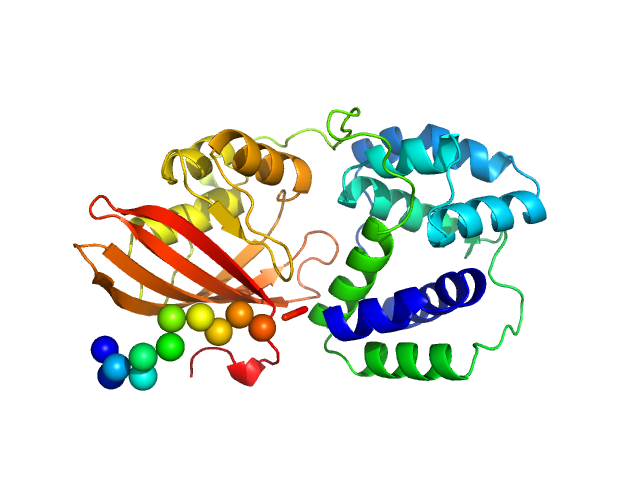
| Sample: |
Orange carotenoid-binding protein monomer, 34 kDa Synechocystis sp. PCC … protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 3% glycerol, pH: 7.6
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Sep 1
|
OCP-FRP protein complex topologies suggest a mechanism for controlling high light tolerance in cyanobacteria.
Nat Commun 9(1):3869 (2018)
Sluchanko NN, Slonimskiy YB, Shirshin EA, Moldenhauer M, Friedrich T, Maksimov EG
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.4 |
nm |
| VolumePorod |
57 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Fluorescence recovery protein dimer, 28 kDa Synechocystis sp. PCC … protein
Orange carotenoid-binding protein monomer, 34 kDa Synechocystis sp. PCC … protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 3% glycerol, pH: 7.6
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Sep 1
|
OCP-FRP protein complex topologies suggest a mechanism for controlling high light tolerance in cyanobacteria.
Nat Commun 9(1):3869 (2018)
Sluchanko NN, Slonimskiy YB, Shirshin EA, Moldenhauer M, Friedrich T, Maksimov EG
|
| RgGuinier |
3.0 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
102 |
nm3 |
|
|
|
|
|
|
|
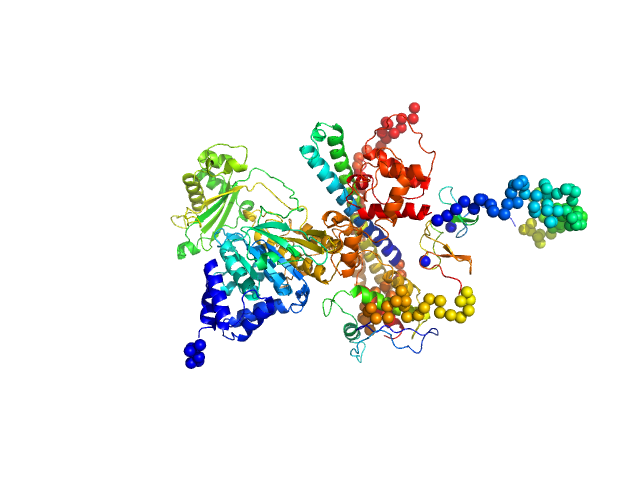
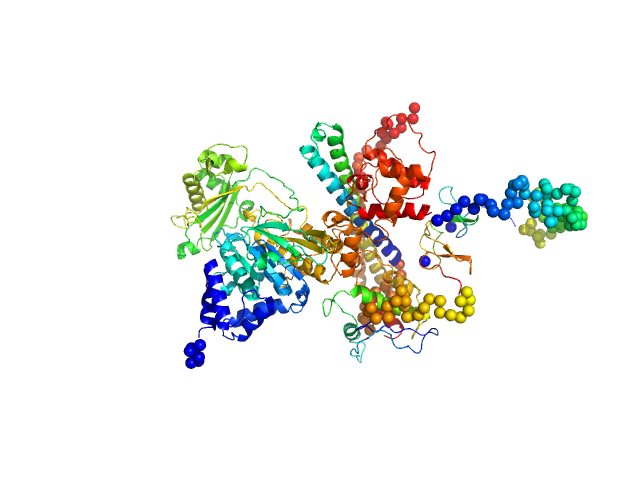
| Sample: |
[F-actin]-monooxygenase MICAL1 (monomer) monomer, 118 kDa Homo sapiens protein
|
| Buffer: |
50 mM sodium phosphate buffer, pH 7.5, 5 % glycerol, 100 mM NaCl, 1 mM EDTA, 1 mM DTT, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Jun 20
|
Human MICAL1: activation by the small GTPase Rab8 and small-angle X-ray scattering studies on the oligomerization state of MICAL1 and its complex with Rab8.
Protein Sci (2018)
Esposito A, Ventura V, Petoukhov MV, Rai A, Svergun DI, Vanoni MA
|
| RgGuinier |
3.7 |
nm |
| Dmax |
12.1 |
nm |
| VolumePorod |
212 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
[F-actin]-monooxygenase MICAL1 (MoChLim) monomer, 85 kDa Homo sapiens protein
|
| Buffer: |
20 mM Hepes/NaOH, pH 7.5, 50 mM NaCl, 2 mM MgCl2, 2 mM DTT, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Jun 20
|
Human MICAL1: activation by the small GTPase Rab8 and small-angle X-ray scattering studies on the oligomerization state of MICAL1 and its complex with Rab8.
Protein Sci (2018)
Esposito A, Ventura V, Petoukhov MV, Rai A, Svergun DI, Vanoni MA
|
| RgGuinier |
4.0 |
nm |
| Dmax |
18.0 |
nm |
| VolumePorod |
145 |
nm3 |
|
|
|
|
|
|
|
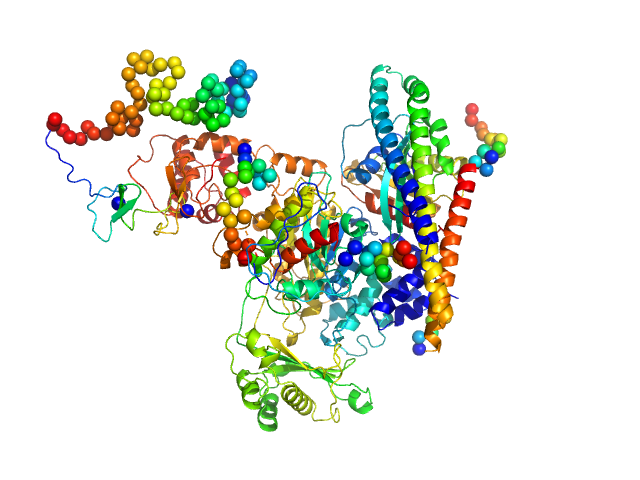
| Sample: |
[F-actin]-monooxygenase MICAL1 (monomer) monomer, 118 kDa Homo sapiens protein
Ras-related protein 8 monomer, 20 kDa protein
|
| Buffer: |
20 mM Hepes/NaOH, pH 7.5, 50 mM NaCl, 2 mM MgCl2, 2 mM DTT, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Jun 20
|
Human MICAL1: activation by the small GTPase Rab8 and small-angle X-ray scattering studies on the oligomerization state of MICAL1 and its complex with Rab8.
Protein Sci (2018)
Esposito A, Ventura V, Petoukhov MV, Rai A, Svergun DI, Vanoni MA
|
| RgGuinier |
3.7 |
nm |
| Dmax |
11.8 |
nm |
| VolumePorod |
234 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
[F-actin]-monooxygenase MICAL1 (MoCh) monomer, 67 kDa Homo sapiens protein
|
| Buffer: |
50 mM sodium phosphate buffer, 5 % glycerol, 100 mM NaCl, 1 mM EDTA, 1 mM DTT, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Jun 6
|
Human MICAL1: activation by the small GTPase Rab8 and small-angle X-ray scattering studies on the oligomerization state of MICAL1 and its complex with Rab8.
Protein Sci (2018)
Esposito A, Ventura V, Petoukhov MV, Rai A, Svergun DI, Vanoni MA
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
100 |
nm3 |
|
|
|
|
|
|
|
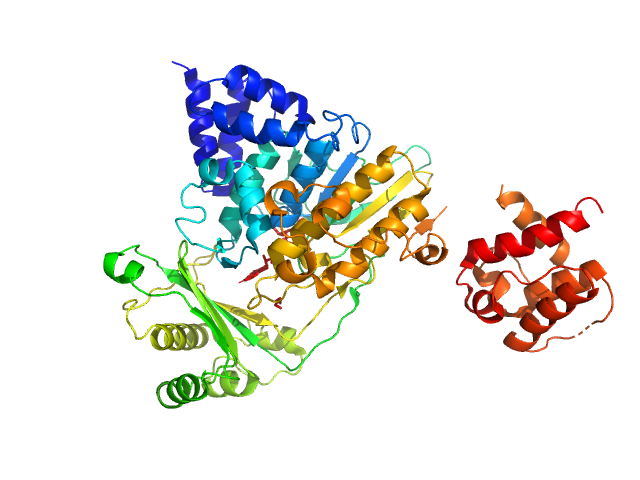
| Sample: |
Neural/ectodermal development factor IMP-L2 dimer, 60 kDa Drosophila melanogaster protein
|
| Buffer: |
phosphate buffered saline, pH: 7.4
|
| Experiment: |
SAXS
data collected at ID14-3, ESRF on 2011 Nov 20
|
Structures of insect Imp-L2 suggest an alternative strategy for regulating the bioavailability of insulin-like hormones.
Nat Commun 9(1):3860 (2018)
Roed NK, Viola CM, Kristensen O, Schluckebier G, Norrman M, Sajid W, Wade JD, Andersen AS, Kristensen C, Ganderton TR, Turkenburg JP, De Meyts P, Brzozowski AM
|
| RgGuinier |
3.1 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
93 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Insulin-like peptide 5 monomer, 5 kDa Drosophila melanogaster protein
Neural/ectodermal development factor IMP-L2 monomer, 30 kDa Drosophila melanogaster protein
|
| Buffer: |
phosphate buffered saline, pH: 7.4
|
| Experiment: |
SAXS
data collected at ID14-3, ESRF on 2011 Nov 20
|
Structures of insect Imp-L2 suggest an alternative strategy for regulating the bioavailability of insulin-like hormones.
Nat Commun 9(1):3860 (2018)
Roed NK, Viola CM, Kristensen O, Schluckebier G, Norrman M, Sajid W, Wade JD, Andersen AS, Kristensen C, Ganderton TR, Turkenburg JP, De Meyts P, Brzozowski AM
|
| RgGuinier |
2.6 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
55 |
nm3 |
|
|