|
|
|
|
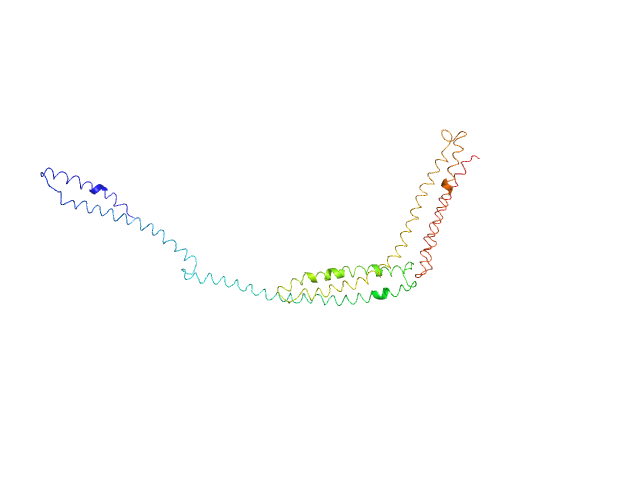
|
| Sample: |
R1-3 human dystrophin fragment monomer, 39 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-d11, 150 mM NaCl, 0.1 mM EDTA-d16, in 100% D2O, pD 7.5, pH: 7.1
|
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2016 Nov 7
|
Human Dystrophin Structural Changes upon Binding to Anionic Membrane Lipids.
Biophys J 115(7):1231-1239 (2018)
Dos Santos Morais R, Delalande O, Pérez J, Mias-Lucquin D, Lagarrigue M, Martel A, Molza AE, Chéron A, Raguénès-Nicol C, Chenuel T, Bondon A, Appavou MS, Le Rumeur E, Combet S, Hubert JF
|
| RgGuinier |
6.2 |
nm |
| Dmax |
24.8 |
nm |
| VolumePorod |
100 |
nm3 |
|
|
|
|
|
|
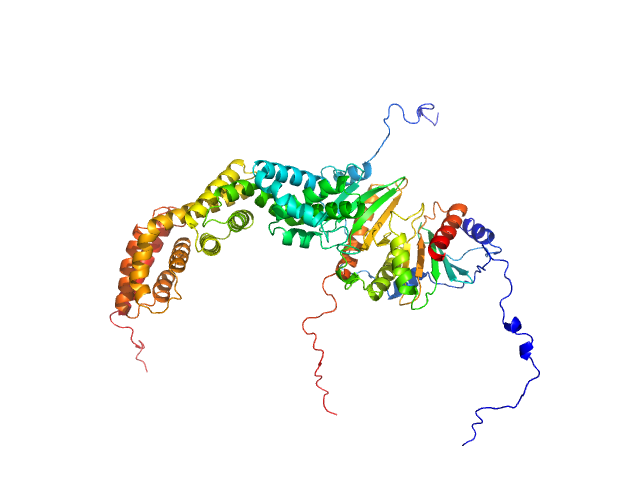
|
| Sample: |
Type III secretion protein HrpG , 19 kDa Erwinia amylovora protein
Type III secretion protein HrpV , 13 kDa Erwinia amylovora protein
Hypersensitivity response secretion protein HrpJ , 42 kDa Erwinia amylovora protein
|
| Buffer: |
20mM Tris-Cl, pH 8.0, 100mM NaCl, 2mM DTT, 0.5mM EDTA, pH: 8
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2016 Apr 3
|
Migration of Type III Secretion System Transcriptional Regulators Links Gene Expression to Secretion.
MBio 9(4) (2018)
Charova SN, Gazi AD, Mylonas E, Pozidis C, Sabarit B, Anagnostou D, Psatha K, Aivaliotis M, Beuzon CR, Panopoulos NJ, Kokkinidis M
|
| RgGuinier |
4.0 |
nm |
| Dmax |
16.0 |
nm |
|
|
|
|
|
|

|
| Sample: |
Fusion protein of LSm and MyoX-coil , 1066 kDa Artificial protein protein
|
| Buffer: |
25 mM HEPES, 100 mM NaCl, 1 mM EDTA, 5% glycerol, pH: 8
|
| Experiment: |
SAXS
data collected at BL-6A, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2017 Nov 11
|
Design of Hollow Protein Nanoparticles with Modifiable Interior and Exterior Surfaces.
Angew Chem Int Ed Engl (2018)
Kawakami N, Kondo H, Matsuzawa Y, Hayasaka K, Nasu E, Sasahara K, Arai R, Miyamoto K
|
| RgGuinier |
9.4 |
nm |
| Dmax |
23.0 |
nm |
|
|
|
|
|
|
|
| Sample: |
Ovalbumin monomer, 43 kDa Gallus gallus protein
|
| Buffer: |
Water, pH: 7
|
| Experiment: |
SAXS
data collected at Bruker Nonius FR591, University of Pennslyvania on 2013 Jun 27
|
The Proof Is in the Pidan: Generalizing Proteins as Patchy Particles.
ACS Cent Sci 4(7):840-853 (2018)
Cai J, Sweeney AM
|
|
|
|
|
|
|
|
| Sample: |
Ovalbumin (common quail) monomer, 42 kDa Coturnix coturnix protein
|
| Buffer: |
Water, pH: 7
|
| Experiment: |
SAXS
data collected at Bruker Nonius FR591, University of Pennslyvania on 2013 Jun 27
|
The Proof Is in the Pidan: Generalizing Proteins as Patchy Particles.
ACS Cent Sci 4(7):840-853 (2018)
Cai J, Sweeney AM
|
|
|
|
|
|
|
|
| Sample: |
Ovalbumin monomer, 43 kDa Gallus gallus protein
|
| Buffer: |
Water, pH: 7
|
| Experiment: |
SAXS
data collected at Bruker Nonius FR591, University of Pennslyvania on 2013 Jun 27
|
The Proof Is in the Pidan: Generalizing Proteins as Patchy Particles.
ACS Cent Sci 4(7):840-853 (2018)
Cai J, Sweeney AM
|
|
|
|
|
|
|

|
| Sample: |
Ovalbumin monomer, 43 kDa Gallus gallus protein
|
| Buffer: |
0.16 M NaOH, pH: 13.2
|
| Experiment: |
SAXS
data collected at X9A, National Synchrotron Light Source (NSLS) on 2014 Feb 21
|
The Proof Is in the Pidan: Generalizing Proteins as Patchy Particles.
ACS Cent Sci 4(7):840-853 (2018)
Cai J, Sweeney AM
|
|
|
|
|
|
|
|
| Sample: |
Anti-CD32b Antibody Clone 6G08 Antibody Binding Fragment monomer, 46 kDa Homo sapiens protein
|
| Buffer: |
50mM HEPES, 150mM KCl, pH: 7.5
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Dec 5
|
Evaluating Anti-CD32b F(ab) Conformation Using Molecular Dynamics and Small-Angle X-Ray Scattering.
Biophys J 115(2):289-299 (2018)
Sutton EJ, Bradshaw RT, Orr CM, Frendéus B, Larsson G, Teige I, Cragg MS, Tews I, Essex JW
|
| RgGuinier |
2.6 |
nm |
| Dmax |
7.3 |
nm |
| VolumePorod |
68 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Small glutamine-rich tetratricopeptide repeat-containing protein alpha full length dimer, 68 kDa Homo sapiens protein
|
| Buffer: |
10 mM potassium phosphate, 100 mM NaCl, pH: 6
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jun 5
|
Structural complexity of the co-chaperone SGTA: a conserved C-terminal region is implicated in dimerization and substrate quality control.
BMC Biol 16(1):76 (2018)
Martínez-Lumbreras S, Krysztofinska EM, Thapaliya A, Spilotros A, Matak-Vinkovic D, Salvadori E, Roboti P, Nyathi Y, Muench JH, Roessler MM, Svergun DI, High S, Isaacson RL
|
|
|
|
|
|
|
|
| Sample: |
Small glutamine-rich tetratricopeptide repeat-containing protein alpha Nterminal-TPR domains dimer, 47 kDa Homo sapiens protein
|
| Buffer: |
10 mM potassium phosphate, 100 mM NaCl, pH: 6
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jun 5
|
Structural complexity of the co-chaperone SGTA: a conserved C-terminal region is implicated in dimerization and substrate quality control.
BMC Biol 16(1):76 (2018)
Martínez-Lumbreras S, Krysztofinska EM, Thapaliya A, Spilotros A, Matak-Vinkovic D, Salvadori E, Roboti P, Nyathi Y, Muench JH, Roessler MM, Svergun DI, High S, Isaacson RL
|
|
|