|
|
|
|
|
| Sample: |
Aureochrome 1a bZIP-LOV module dimer, 57 kDa Phaeodactylum tricornutum protein
|
| Buffer: |
10 mM Tris 300 mM NaCl, pH: 8
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Nov 6
|
Allosteric communication between DNA-binding and light-responsive domains of diatom class I aureochromes.
Nucleic Acids Res 44(12):5957-70 (2016)
Banerjee A, Herman E, Serif M, Maestre-Reyna M, Hepp S, Pokorny R, Kroth PG, Essen LO, Kottke T
|
| RgGuinier |
3.9 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
121 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Aureochrome 1a bZIP-LOV module dimer, 57 kDa Phaeodactylum tricornutum protein
|
| Buffer: |
10 mM Tris 300 mM NaCl, pH: 8
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Nov 6
|
Allosteric communication between DNA-binding and light-responsive domains of diatom class I aureochromes.
Nucleic Acids Res 44(12):5957-70 (2016)
Banerjee A, Herman E, Serif M, Maestre-Reyna M, Hepp S, Pokorny R, Kroth PG, Essen LO, Kottke T
|
| RgGuinier |
3.8 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
117 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
short-crRNA: CRISPR/Cas Type I-F Cascade component monomer, 14 kDa Shewanella putrefaciens RNA
Cas6f: CRISPR/Cas Type I-F Cascade component (CRISPR-associated protein, Csy4 family) monomer, 21 kDa Shewanella putrefaciens protein
Trimeric Cas7fv: CRISPR/Cas Type I-F Cascade component (Uncharacterized protein, Sputcn32_1821) trimer, 112 kDa Shewanella putrefaciens protein
Cas5fv: CRISPR/Cas Type I-F Cascade component (Uncharacterized protein, Sputcn32_1822) monomer, 38 kDa Shewanella putrefaciens protein
|
| Buffer: |
50 mM HEPES 150 mM NaCl 1mM DTT 1mM EDTA, pH: 7
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Jun 27
|
Modulating the Cascade architecture of a minimal Type I-F CRISPR-Cas system.
Nucleic Acids Res 44(12):5872-82 (2016)
Gleditzsch D, Müller-Esparza H, Pausch P, Sharma K, Dwarakanath S, Urlaub H, Bange G, Randau L
|
| RgGuinier |
4.1 |
nm |
| Dmax |
14.2 |
nm |
|
|
|
|
|
|
|
| Sample: |
Cas6f: CRISPR/Cas Type I-F Cascade component (CRISPR-associated protein, Csy4 family) monomer, 21 kDa Shewanella putrefaciens protein
Cas5fv: CRISPR/Cas Type I-F Cascade component (Uncharacterized protein, Sputcn32_1822) monomer, 38 kDa Shewanella putrefaciens protein
Hexameric Cas7fv: CRISPR/Cas Type I-F Cascade component (Uncharacterized protein, Sputcn32_1821) hexamer, 223 kDa Shewanella putrefaciens protein
wildtype-crRNA: CRISPR/Cas Type I-F Cascade component monomer, 19 kDa RNA
|
| Buffer: |
50 mM HEPES 150 mM NaCl 1mM DTT 1mM EDTA, pH: 7
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Jun 27
|
Modulating the Cascade architecture of a minimal Type I-F CRISPR-Cas system.
Nucleic Acids Res 44(12):5872-82 (2016)
Gleditzsch D, Müller-Esparza H, Pausch P, Sharma K, Dwarakanath S, Urlaub H, Bange G, Randau L
|
| RgGuinier |
5.4 |
nm |
| Dmax |
18.4 |
nm |
|
|
|
|
|
|

|
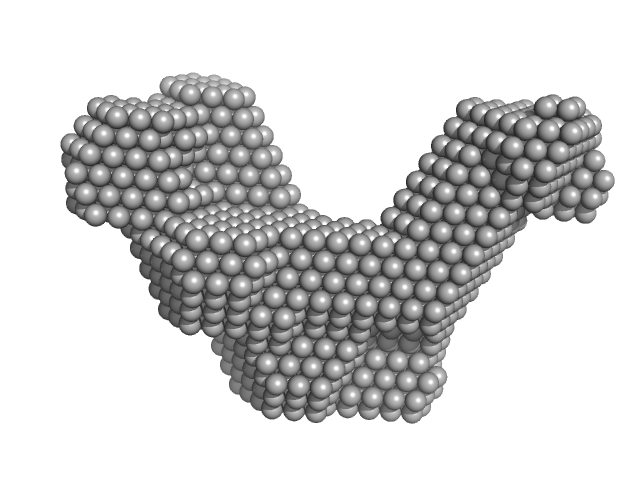
| Sample: |
Cas6f: CRISPR/Cas Type I-F Cascade component (CRISPR-associated protein, Csy4 family) monomer, 21 kDa Shewanella putrefaciens protein
Cas5fv: CRISPR/Cas Type I-F Cascade component (Uncharacterized protein, Sputcn32_1822) monomer, 38 kDa Shewanella putrefaciens protein
Nonameric Cas7fv: CRISPR/Cas Type I-F Cascade component (Uncharacterized protein, Sputcn32_1821) nonamer, 335 kDa Shewanella putrefaciens protein
long-crRNA: CRISPR/Cas Type I-F Cascade component monomer, 25 kDa RNA
|
| Buffer: |
50 mM HEPES 150 mM NaCl 1mM DTT 1mM EDTA, pH: 7
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Jan 30
|
Modulating the Cascade architecture of a minimal Type I-F CRISPR-Cas system.
Nucleic Acids Res 44(12):5872-82 (2016)
Gleditzsch D, Müller-Esparza H, Pausch P, Sharma K, Dwarakanath S, Urlaub H, Bange G, Randau L
|
| RgGuinier |
6.5 |
nm |
| Dmax |
21.6 |
nm |
|
|
|
|
|
|
|
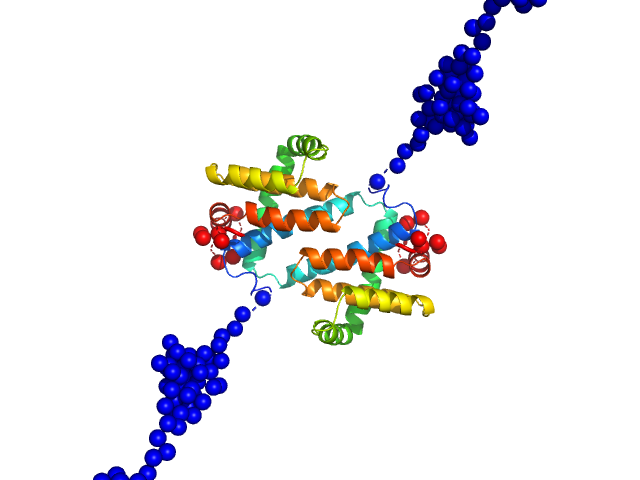
| Sample: |
MVA F1L antiapoptotic Bcl-2 viral protein dimer, 51 kDa Vaccinia virus protein
|
| Buffer: |
25 mM HEPES 150 mM NaCl 5 mM DTT, pH: 7.5
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2016 Apr 15
|
The N Terminus of the Vaccinia Virus Protein F1L Is an Intrinsically Unstructured Region That Is Not Involved in Apoptosis Regulation.
J Biol Chem 291(28):14600-8 (2016)
Caria S, Marshall B, Burton RL, Campbell S, Pantaki-Eimany D, Hawkins CJ, Barry M, Kvansakul M
|
| RgGuinier |
3.4 |
nm |
| Dmax |
16.2 |
nm |
| VolumePorod |
74 |
nm3 |
|
|
|
|
|
|
|
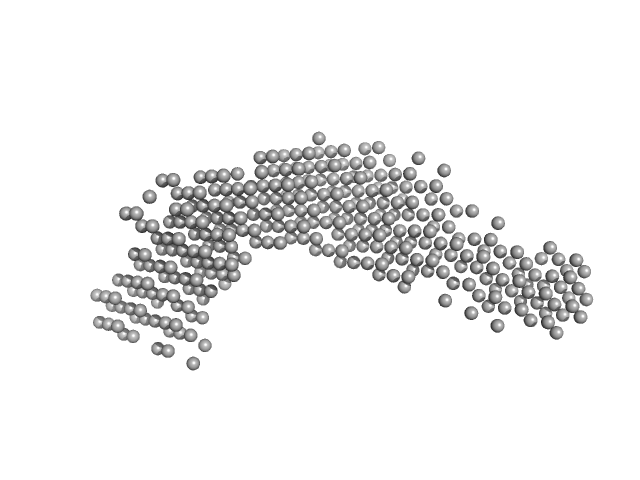
| Sample: |
Stress-induced protein sti1 monomer, 62 kDa Leishmania braziliensis protein
|
| Buffer: |
25 mM Tris 100 mM NaCl 1 mM EDTA 1 mM β-mercaptoethanol, pH: 7.5
|
| Experiment: |
SAXS
data collected at SAXS1 Beamline, Brazilian Synchrotron Light Laboratory on 2016 Feb 21
|
Low sequence identity but high structural and functional conservation: The case of Hsp70/Hsp90 organizing protein (Hop/Sti1) of Leishmania braziliensis.
Arch Biochem Biophys 600:12-22 (2016)
Batista FAH, Seraphim TV, Santos CA, Gonzaga MR, Barbosa LRS, Ramos CHI, Borges JC
|
| RgGuinier |
4.5 |
nm |
| Dmax |
18.0 |
nm |
| VolumePorod |
94 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Stress-induced protein sti1 (Hop TPR2A-TPR2B-DP2 construct) monomer, 44 kDa Leishmania braziliensis protein
|
| Buffer: |
25 mM Tris 100 mM NaCl 1 mM EDTA 1 mM β-mercaptoethanol, pH: 7.5
|
| Experiment: |
SAXS
data collected at SAXS2 Beamline, Brazilian Synchrotron Light Laboratory on 2016 Feb 21
|
Low sequence identity but high structural and functional conservation: The case of Hsp70/Hsp90 organizing protein (Hop/Sti1) of Leishmania braziliensis.
Arch Biochem Biophys 600:12-22 (2016)
Batista FAH, Seraphim TV, Santos CA, Gonzaga MR, Barbosa LRS, Ramos CHI, Borges JC
|
| RgGuinier |
3.8 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
65 |
nm3 |
|
|
|
|
|
|
|
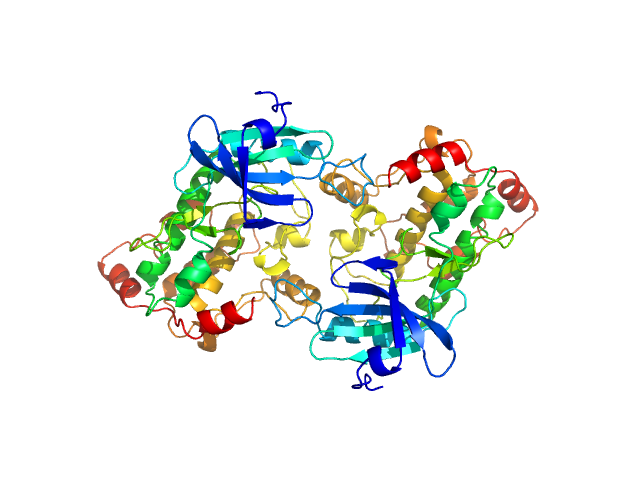
| Sample: |
Death associated protein kinase wild-type , 37 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES 250 mM NaCl 5mM CaCl2 0.25 mM TCEP 5% (v/v) glycerol, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Dec 18
|
Death-Associated Protein Kinase Activity Is Regulated by Coupled Calcium/Calmodulin Binding to Two Distinct Sites.
Structure 24(6):851-61 (2016)
Simon B, Huart AS, Temmerman K, Vahokoski J, Mertens HD, Komadina D, Hoffmann JE, Yumerefendi H, Svergun DI, Kursula P, Schultz C, McCarthy AA, Hart DJ, Wilmanns M
|
| RgGuinier |
2.7 |
nm |
| Dmax |
8.1 |
nm |
| VolumePorod |
101 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Death associated protein kinase (D220K mutant) , 37 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES 250 mM NaCl 5mM CaCl2 0.25 mM TCEP 5% (v/v) glycerol, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Dec 18
|
Death-Associated Protein Kinase Activity Is Regulated by Coupled Calcium/Calmodulin Binding to Two Distinct Sites.
Structure 24(6):851-61 (2016)
Simon B, Huart AS, Temmerman K, Vahokoski J, Mertens HD, Komadina D, Hoffmann JE, Yumerefendi H, Svergun DI, Kursula P, Schultz C, McCarthy AA, Hart DJ, Wilmanns M
|
| RgGuinier |
2.5 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
89 |
nm3 |
|
|