|
|
|
|
|
| Sample: |
Plakin domain of Human Desmoplakin monomer, 99 kDa Homo sapiens protein
|
| Buffer: |
20 mM Sodium Phosphate 150 mM NaCl 5% glycerol 3 mM DTT, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Sep 25
|
The Structure of the Plakin Domain of Plectin Reveals an Extended Rod-like Shape.
J Biol Chem 291(36):18643-62 (2016)
Ortega E, Manso JA, Buey RM, Carballido AM, Carabias A, Sonnenberg A, de Pereda JM
|
| RgGuinier |
7.5 |
nm |
| Dmax |
35.0 |
nm |
| VolumePorod |
136 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Functional binding region (187-385) of the pneumococcal serine-rich repeat protein monomer, 22 kDa Streptococcus pneumoniae protein
|
| Buffer: |
PBS 5 % Glycerol, pH: 7.4
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2013 Feb 27
|
The BR domain of PsrP interacts with extracellular DNA to promote bacterial aggregation; structural insights into pneumococcal biofilm formation.
Sci Rep 6:32371 (2016)
Schulte T, Mikaelsson C, Beaussart A, Kikhney A, Deshmukh M, Wolniak S, Pathak A, Ebel C, Löfling J, Fogolari F, Henriques-Normark B, Dufrêne YF, Svergun D, Nygren PÅ, Achour A
|
| RgGuinier |
2.0 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
38 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Functional binding region (120-395) of the pneumococcal serine-rich repeat protein monomer, 30 kDa Streptococcus pneumoniae protein
|
| Buffer: |
PBS 5 % Glycerol, pH: 7.4
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2013 Feb 27
|
The BR domain of PsrP interacts with extracellular DNA to promote bacterial aggregation; structural insights into pneumococcal biofilm formation.
Sci Rep 6:32371 (2016)
Schulte T, Mikaelsson C, Beaussart A, Kikhney A, Deshmukh M, Wolniak S, Pathak A, Ebel C, Löfling J, Fogolari F, Henriques-Normark B, Dufrêne YF, Svergun D, Nygren PÅ, Achour A
|
| RgGuinier |
2.9 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
41 |
nm3 |
|
|
|
|
|
|
|
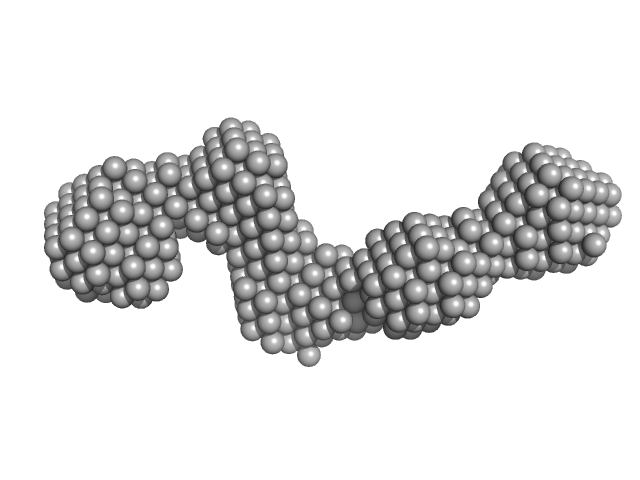
| Sample: |
Pre-B-cell leukemia transcription factor 1 monomer, 34 kDa Homo sapiens protein
Homeobox protein PKNOX1 monomer, 38 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-HCl 150 mM NaCl 5% glycerol 1 mM DTT, pH: 7.4
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Dec 6
|
The flexibility of a homeodomain transcription factor heterodimer and its allosteric regulation by DNA binding.
FEBS J 283(16):3134-54 (2016)
Mathiasen L, Valentini E, Boivin S, Cattaneo A, Blasi F, Svergun DI, Bruckmann C
|
| RgGuinier |
5.8 |
nm |
| Dmax |
19.0 |
nm |
| VolumePorod |
143 |
nm3 |
|
|
|
|
|
|
|
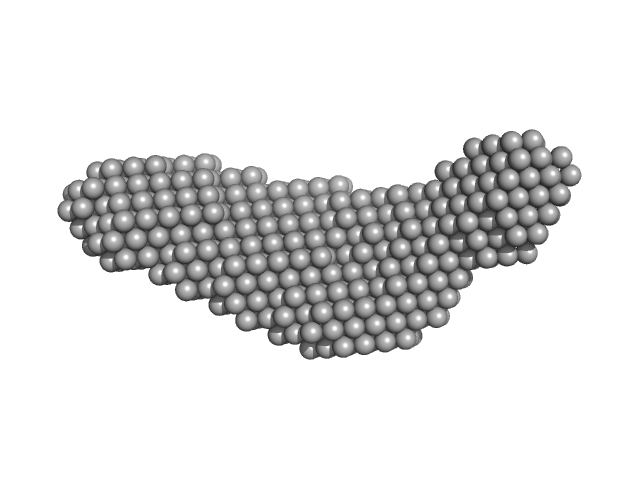
| Sample: |
Pre-B-cell leukemia transcription factor 1 monomer, 34 kDa Homo sapiens protein
Homeobox protein PKNOX1 monomer, 38 kDa Homo sapiens protein
DNA 22mer monomer, 12 kDa DNA
|
| Buffer: |
20 mM Tris-HCl 150 mM NaCl 5% glycerol 1 mM DTT, pH: 7.4
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Dec 6
|
The flexibility of a homeodomain transcription factor heterodimer and its allosteric regulation by DNA binding.
FEBS J 283(16):3134-54 (2016)
Mathiasen L, Valentini E, Boivin S, Cattaneo A, Blasi F, Svergun DI, Bruckmann C
|
| RgGuinier |
4.8 |
nm |
| Dmax |
16.3 |
nm |
| VolumePorod |
173 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
DNA 22mer monomer, 12 kDa DNA
|
| Buffer: |
20 mM Tris-HCl 150 mM NaCl 5% glycerol 1 mM DTT, pH: 7.4
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Dec 6
|
The flexibility of a homeodomain transcription factor heterodimer and its allosteric regulation by DNA binding.
FEBS J 283(16):3134-54 (2016)
Mathiasen L, Valentini E, Boivin S, Cattaneo A, Blasi F, Svergun DI, Bruckmann C
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
19 |
nm3 |
|
|
|
|
|
|
|
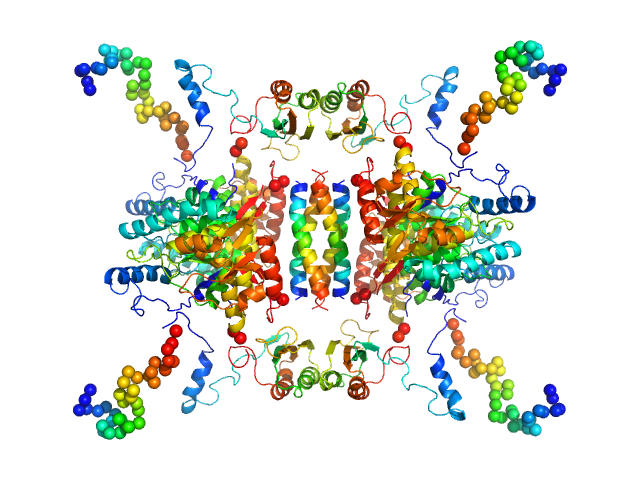
| Sample: |
Tyrosine hydroxylase, isoform 1 tetramer, 222 kDa Homo sapiens protein
|
| Buffer: |
20 mM Na-HEPES 200 mM NaCl, pH: 7
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jun 14
|
Stable preparations of tyrosine hydroxylase provide the solution structure of the full-length enzyme.
Sci Rep 6:30390 (2016)
Bezem MT, Baumann A, Skjærven L, Meyer R, Kursula P, Martinez A, Flydal MI
|
| RgGuinier |
4.7 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
520 |
nm3 |
|
|
|
|
|
|
|
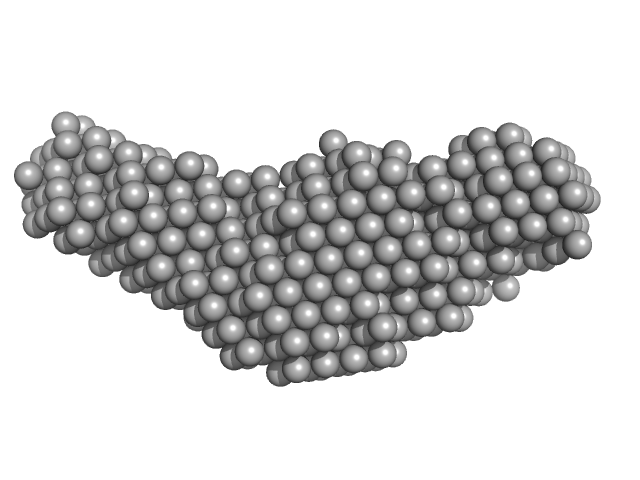
| Sample: |
Aureobox dsDNA monomer, 13 kDa synthetic construct DNA
|
| Buffer: |
50 mM Tris 50 mM boric acid 1 mM EDTA, pH: 8
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Nov 6
|
Allosteric communication between DNA-binding and light-responsive domains of diatom class I aureochromes.
Nucleic Acids Res 44(12):5957-70 (2016)
Banerjee A, Herman E, Serif M, Maestre-Reyna M, Hepp S, Pokorny R, Kroth PG, Essen LO, Kottke T
|
| RgGuinier |
2.0 |
nm |
| Dmax |
8.6 |
nm |
| VolumePorod |
18 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Aureochrome 1a bZIP-LOV module dimer, 57 kDa Phaeodactylum tricornutum protein
|
| Buffer: |
50 mM Tris 50 mM boric acid 1 mM EDTA, pH: 8
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Nov 6
|
Allosteric communication between DNA-binding and light-responsive domains of diatom class I aureochromes.
Nucleic Acids Res 44(12):5957-70 (2016)
Banerjee A, Herman E, Serif M, Maestre-Reyna M, Hepp S, Pokorny R, Kroth PG, Essen LO, Kottke T
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.6 |
nm |
| VolumePorod |
115 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Aureobox dsDNA monomer, 13 kDa synthetic construct DNA
Aureochrome 1a bZIP-LOV module dimer, 57 kDa Phaeodactylum tricornutum protein
|
| Buffer: |
50 mM Tris 50 mM boric acid 1 mM EDTA, pH: 8
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Nov 6
|
Allosteric communication between DNA-binding and light-responsive domains of diatom class I aureochromes.
Nucleic Acids Res 44(12):5957-70 (2016)
Banerjee A, Herman E, Serif M, Maestre-Reyna M, Hepp S, Pokorny R, Kroth PG, Essen LO, Kottke T
|
| RgGuinier |
4.5 |
nm |
| Dmax |
16.7 |
nm |
| VolumePorod |
97 |
nm3 |
|
|