|
|
|
|
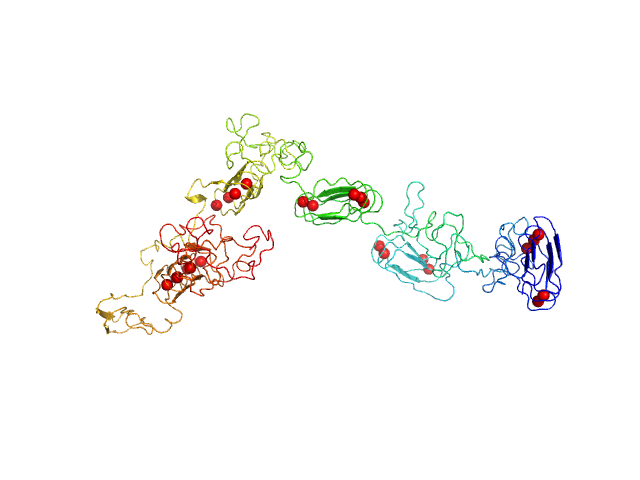
|
| Sample: |
RD domain of B. Pertussis Adenylate Cyclase Toxin (CyaA) monomer, 73 kDa Bordetella pertussis protein
|
| Buffer: |
20 mM Hepes, 150 mM NaCl, 2 mM DTT, 4 mM CaCl2, pH: 7.5
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2012 May 31
|
Structural models of intrinsically disordered and calcium-bound folded states of a protein adapted for secretion.
Sci Rep 5:14223 (2015)
O'Brien DP, Hernandez B, Durand D, Hourdel V, Sotomayor-Pérez AC, Vachette P, Ghomi M, Chamot-Rooke J, Ladant D, Brier S, Chenal A
|
| RgGuinier |
4.4 |
nm |
| Dmax |
15.5 |
nm |
| VolumePorod |
89 |
nm3 |
|
|
|
|
|
|
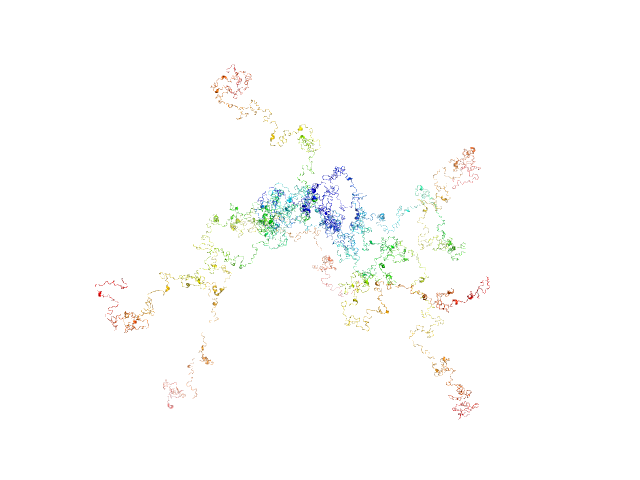
|
| Sample: |
RD domain of B. Pertussis Adenylate Cyclase Toxin (CyaA) monomer, 73 kDa Bordetella pertussis protein
|
| Buffer: |
20 mM Hepes, 150 mM NaCl, 2 mM DTT, pH: 7.5
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2012 May 31
|
Structural models of intrinsically disordered and calcium-bound folded states of a protein adapted for secretion.
Sci Rep 5:14223 (2015)
O'Brien DP, Hernandez B, Durand D, Hourdel V, Sotomayor-Pérez AC, Vachette P, Ghomi M, Chamot-Rooke J, Ladant D, Brier S, Chenal A
|
| RgGuinier |
8.3 |
nm |
| Dmax |
33.0 |
nm |
|
|
|
|
|
|
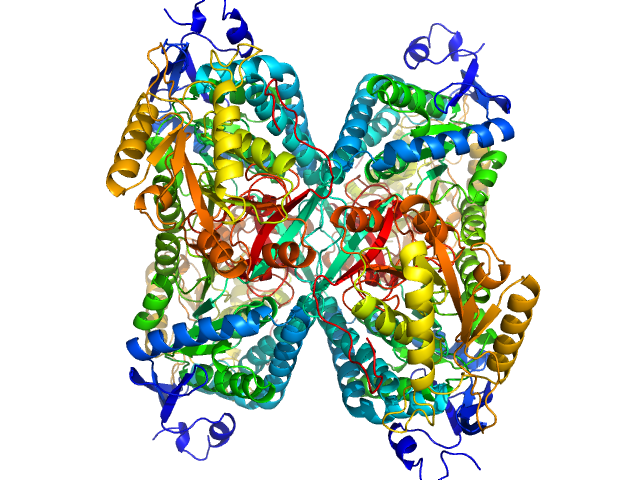
|
| Sample: |
Aldehyde dehydrogenase 7A1 (Alpha-aminoadipic semialdehyde dehydrogenase) tetramer, 222 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris, 5% glycerol, 0.5 mM tris(3-hydroxypropyl)phosphine, 50 mM NaCl, pH: 7.8
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2014 Mar 9
|
Structural Basis of Substrate Recognition by Aldehyde Dehydrogenase 7A1.
Biochemistry 54(35):5513-22 (2015)
Luo M, Tanner JJ
|
| RgGuinier |
3.8 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
270 |
nm3 |
|
|
|
|
|
|
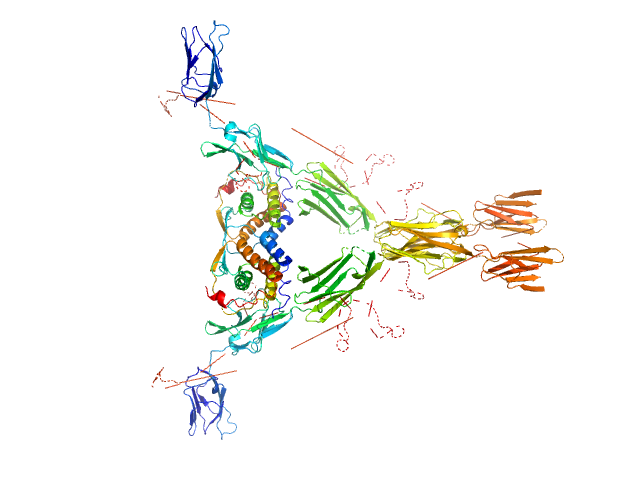
|
| Sample: |
Macrophage colony-stimulating factor 1 dimer, 35 kDa Homo sapiens protein
Macrophage colony-stimulating factor 1 receptor dimer, 107 kDa Homo sapiens protein
|
| Buffer: |
50 mM NaH2PO4, 100 m, pH: 7.4
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Mar 13
|
Structure and Assembly Mechanism of the Signaling Complex Mediated by Human CSF-1.
Structure 23(9):1621-1631 (2015)
Felix J, De Munck S, Verstraete K, Meuris L, Callewaert N, Elegheert J, Savvides SN
|
| RgGuinier |
5.7 |
nm |
| Dmax |
17.9 |
nm |
| VolumePorod |
299 |
nm3 |
|
|
|
|
|
|
|
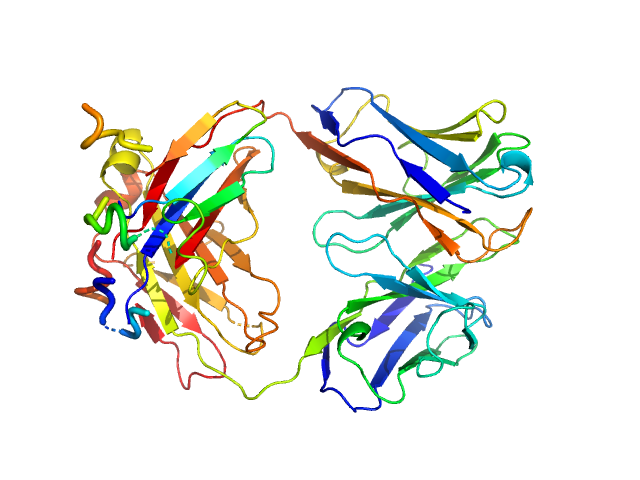
| Sample: |
anti-TG2 antibody (679 14 E06) monomer, 48 kDa protein
|
| Buffer: |
20 mM Tris 150mM NaCl 1mM EDTA, pH: 7.2
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jan 17
|
Structural Basis for Antigen Recognition by Transglutaminase 2-specific Autoantibodies in Celiac Disease.
J Biol Chem 290(35):21365-75 (2015)
Chen X, Hnida K, Graewert MA, Andersen JT, Iversen R, Tuukkanen A, Svergun D, Sollid LM
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.1 |
nm |
| VolumePorod |
58 |
nm3 |
|
|
|
|
|
|
|
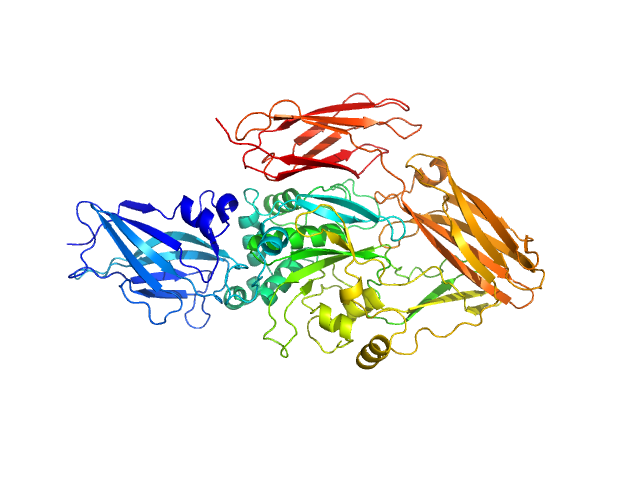
| Sample: |
transglutaminase 2 monomer, 79 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris 150mM NaCl 1mM EDTA, pH: 7.2
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jan 17
|
Structural Basis for Antigen Recognition by Transglutaminase 2-specific Autoantibodies in Celiac Disease.
J Biol Chem 290(35):21365-75 (2015)
Chen X, Hnida K, Graewert MA, Andersen JT, Iversen R, Tuukkanen A, Svergun D, Sollid LM
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
117 |
nm3 |
|
|
|
|
|
|

|
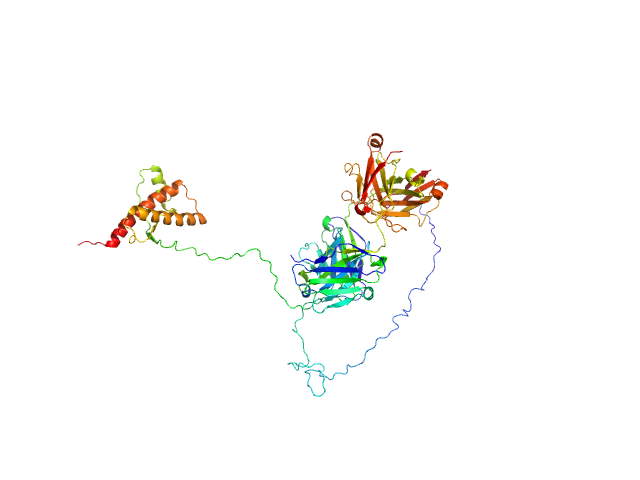
| Sample: |
anti-TG2 antibody (679 14 E06) monomer, 48 kDa protein
transglutaminase 2 monomer, 79 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris 150mM NaCl 1mM EDTA, pH: 7.2
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jan 17
|
Structural Basis for Antigen Recognition by Transglutaminase 2-specific Autoantibodies in Celiac Disease.
J Biol Chem 290(35):21365-75 (2015)
Chen X, Hnida K, Graewert MA, Andersen JT, Iversen R, Tuukkanen A, Svergun D, Sollid LM
|
| RgGuinier |
4.0 |
nm |
| Dmax |
13.9 |
nm |
| VolumePorod |
168 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Major prion protein monomer, 23 kDa Mus musculus protein
P-Clone Fab, Chimera monomer, 47 kDa Homo sapiens protein
|
| Buffer: |
sodium acetate buffer (20 mM sodium acetate, pH 5.1; 150 mM NaCl), pH: 5.1
|
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2013 Dec 5
|
Prion Protein-Antibody Complexes Characterized by Chromatography-Coupled Small-Angle X-Ray Scattering.
Biophys J 109(4):793-805 (2015)
Carter L, Kim SJ, Schneidman-Duhovny D, Stöhr J, Poncet-Montange G, Weiss TM, Tsuruta H, Prusiner SB, Sali A
|
| RgGuinier |
3.9 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
106 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Hyaluronate binding domain of CD44 antigen monomer, 18 kDa Homo sapiens protein
Single-chain Variable Fragment of Antibody MEM-85 monomer, 29 kDa Mus musculus protein
|
| Buffer: |
PBS, pH: 7.4
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Oct 31
|
Molecular mechanism for the action of the anti-CD44 monoclonal antibody MEM-85.
J Struct Biol 191(2):214-23 (2015)
Škerlová J, Král V, Kachala M, Fábry M, Bumba L, Svergun DI, Tošner Z, Veverka V, Řezáčová P
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.4 |
nm |
| VolumePorod |
57 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Antiapoptotic membrane protein dimer, 39 kDa Deerpox virus W-1170-84 protein
|
| Buffer: |
25 mM HEPES 150 mM NaCl, pH: 7.5
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2013 May 4
|
Structural basis of Deerpox virus-mediated inhibition of apoptosis.
Acta Crystallogr D Biol Crystallogr 71(Pt 8):1593-603 (2015)
Burton DR, Caria S, Marshall B, Barry M, Kvansakul M
|
| RgGuinier |
2.6 |
nm |
| Dmax |
11.1 |
nm |
| VolumePorod |
61 |
nm3 |
|
|