|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDM39_fit1_model1.png)
|
| Sample: |
Iron oxide nanoparticles (NP-P3) encapsulated into brome mosaic virus (BMV) 0, 5000 kDa
|
| Buffer: |
50 mM Tris-HCl, 50 mM NaCl, 10 mM KCl, 5 mM MgCl2, pH: 4.5
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Jun 23
|
Coat Protein-Dependent Behavior of Poly(ethylene glycol) Tails in Iron Oxide Core Virus-like Nanoparticles.
ACS Appl Mater Interfaces 7(22):12089-98 (2015)
Malyutin AG, Cheng H, Sanchez-Felix OR, Carlson K, Stein BD, Konarev PV, Svergun DI, Dragnea B, Bronstein LM
|
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDM49_fit1_model1.png)
|
| Sample: |
Iron oxide nanoparticles (NP-N3) encapsulated into hepatitis B virus (HBV) 0, 5000 kDa
|
| Buffer: |
0.5 M LiCl, 50 mM HEPES, 2 mM DTT, pH 7.5, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Jun 23
|
Coat Protein-Dependent Behavior of Poly(ethylene glycol) Tails in Iron Oxide Core Virus-like Nanoparticles.
ACS Appl Mater Interfaces 7(22):12089-98 (2015)
Malyutin AG, Cheng H, Sanchez-Felix OR, Carlson K, Stein BD, Konarev PV, Svergun DI, Dragnea B, Bronstein LM
|
|
|
|
|
|
|
|
| Sample: |
brome mosaic virus (BMV) monomer, 5000 kDa
|
| Buffer: |
50 mM Tris-HCl, 50 mM NaCl, 10 mM KCl, 5 mM MgCl2, pH: 4.6
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Jun 23
|
Coat Protein-Dependent Behavior of Poly(ethylene glycol) Tails in Iron Oxide Core Virus-like Nanoparticles.
ACS Appl Mater Interfaces 7(22):12089-98 (2015)
Malyutin AG, Cheng H, Sanchez-Felix OR, Carlson K, Stein BD, Konarev PV, Svergun DI, Dragnea B, Bronstein LM
|
|
|
|
|
|
|
|
| Sample: |
Hepatitis B virus (HBV) None,
|
| Buffer: |
0.5 M LiCl, 50 mM HEPES, 2 mM DTT, pH 7.5, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Jun 23
|
Coat Protein-Dependent Behavior of Poly(ethylene glycol) Tails in Iron Oxide Core Virus-like Nanoparticles.
ACS Appl Mater Interfaces 7(22):12089-98 (2015)
Malyutin AG, Cheng H, Sanchez-Felix OR, Carlson K, Stein BD, Konarev PV, Svergun DI, Dragnea B, Bronstein LM
|
|
|
|
|
|
|
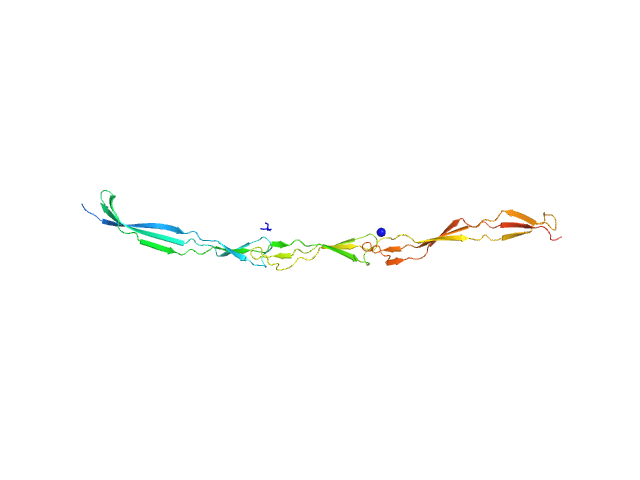
|
| Sample: |
Surface protein G monomer, 24 kDa Staphylococcus aureus protein
|
| Buffer: |
20 mM Tris 200 mM NaCl 1 mM EDTA 20 mM Tris.Cl, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Nov 12
|
Cooperative folding of intrinsically disordered domains drives assembly of a strong elongated protein.
Nat Commun 6:7271 (2015)
Gruszka DT, Whelan F, Farrance OE, Fung HK, Paci E, Jeffries CM, Svergun DI, Baldock C, Baumann CG, Brockwell DJ, Potts JR, Clarke J
|
| RgGuinier |
4.7 |
nm |
| Dmax |
19.0 |
nm |
| VolumePorod |
29 |
nm3 |
|
|
|
|
|
|
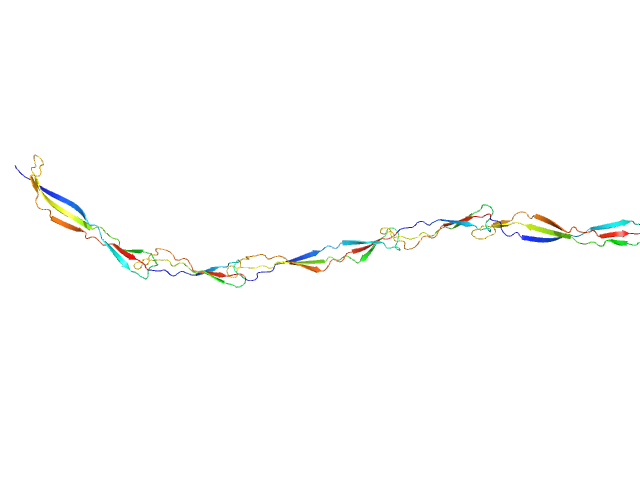
|
| Sample: |
Surface protein G monomer, 39 kDa Staphylococcus aureus protein
|
| Buffer: |
20 mM Tris 200 mM NaCl 1 mM EDTA 20 mM Tris.Cl, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Nov 12
|
Cooperative folding of intrinsically disordered domains drives assembly of a strong elongated protein.
Nat Commun 6:7271 (2015)
Gruszka DT, Whelan F, Farrance OE, Fung HK, Paci E, Jeffries CM, Svergun DI, Baldock C, Baumann CG, Brockwell DJ, Potts JR, Clarke J
|
| RgGuinier |
7.7 |
nm |
| Dmax |
30.5 |
nm |
| VolumePorod |
49 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Surface protein G monomer, 53 kDa Staphylococcus aureus protein
|
| Buffer: |
20 mM Tris 200 mM NaCl 1 mM EDTA 20 mM Tris.Cl, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Nov 12
|
Cooperative folding of intrinsically disordered domains drives assembly of a strong elongated protein.
Nat Commun 6:7271 (2015)
Gruszka DT, Whelan F, Farrance OE, Fung HK, Paci E, Jeffries CM, Svergun DI, Baldock C, Baumann CG, Brockwell DJ, Potts JR, Clarke J
|
| RgGuinier |
9.7 |
nm |
| Dmax |
38.5 |
nm |
| VolumePorod |
58 |
nm3 |
|
|
|
|
|
|
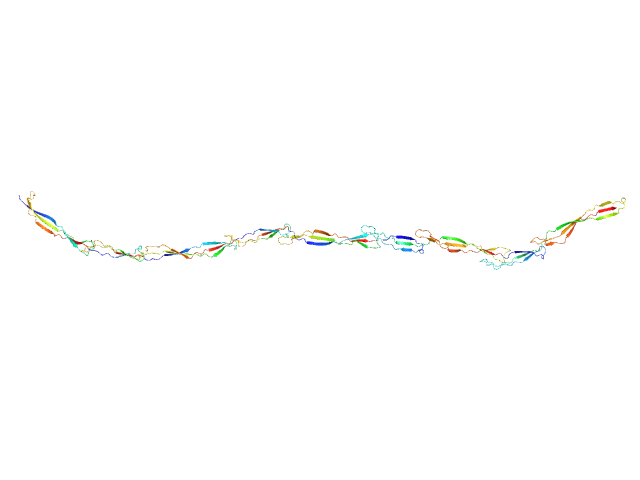
|
| Sample: |
Surface protein G monomer, 67 kDa Staphylococcus aureus protein
|
| Buffer: |
20 mM Tris 200 mM NaCl 1 mM EDTA 20 mM Tris.Cl, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Nov 12
|
Cooperative folding of intrinsically disordered domains drives assembly of a strong elongated protein.
Nat Commun 6:7271 (2015)
Gruszka DT, Whelan F, Farrance OE, Fung HK, Paci E, Jeffries CM, Svergun DI, Baldock C, Baumann CG, Brockwell DJ, Potts JR, Clarke J
|
| RgGuinier |
12.0 |
nm |
| Dmax |
48.0 |
nm |
| VolumePorod |
87 |
nm3 |
|
|
|
|
|
|
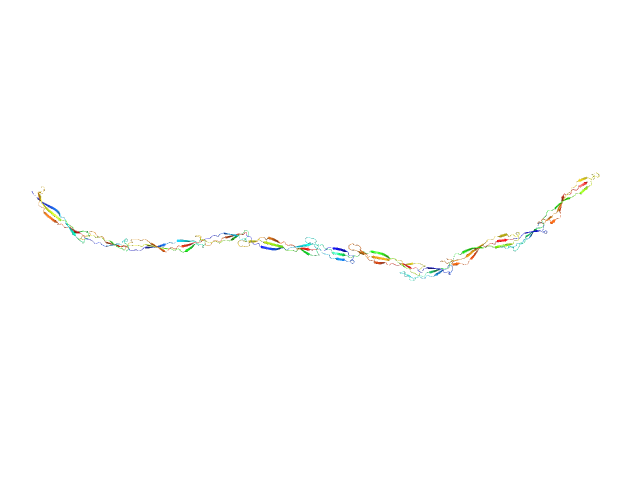
|
| Sample: |
Surface protein G monomer, 81 kDa Staphylococcus aureus protein
|
| Buffer: |
20 mM Tris 200 mM NaCl 1 mM EDTA 20 mM Tris.Cl, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Nov 12
|
Cooperative folding of intrinsically disordered domains drives assembly of a strong elongated protein.
Nat Commun 6:7271 (2015)
Gruszka DT, Whelan F, Farrance OE, Fung HK, Paci E, Jeffries CM, Svergun DI, Baldock C, Baumann CG, Brockwell DJ, Potts JR, Clarke J
|
| RgGuinier |
14.1 |
nm |
| Dmax |
57.0 |
nm |
| VolumePorod |
89 |
nm3 |
|
|
|
|
|
|
|
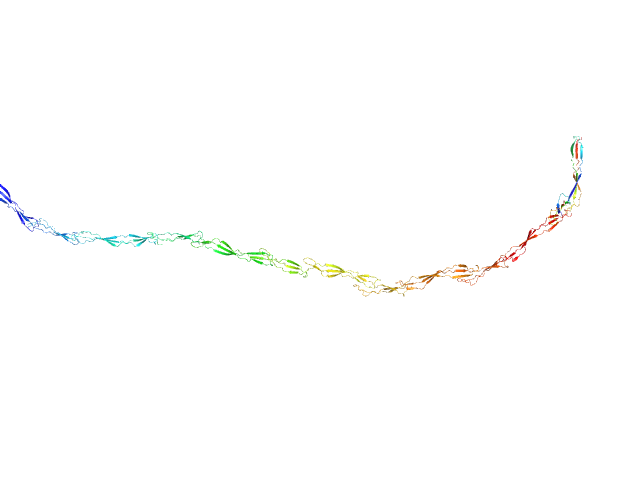
| Sample: |
Surface protein G monomer, 95 kDa Staphylococcus aureus protein
|
| Buffer: |
20 mM Tris 200 mM NaCl 1 mM EDTA 20 mM Tris.Cl, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Nov 12
|
Cooperative folding of intrinsically disordered domains drives assembly of a strong elongated protein.
Nat Commun 6:7271 (2015)
Gruszka DT, Whelan F, Farrance OE, Fung HK, Paci E, Jeffries CM, Svergun DI, Baldock C, Baumann CG, Brockwell DJ, Potts JR, Clarke J
|
| RgGuinier |
15.9 |
nm |
| Dmax |
63.0 |
nm |
| VolumePorod |
122 |
nm3 |
|
|